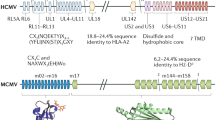
Abstract
Depending on ethnicity and on social conditions, between 40 and 90 % of the population is infected with human cytomegalovirus (HCMV). In immunocompetent patients, the virus may cause an acute disease and then revert to a state of latency, which enables its coexistence with the human host. However, in cases of immunosuppression or in neonatal infections, HCMV can cause serious long-lasting illnesses. HCMV has developed multiple mechanisms in order to escape its elimination by the immune system, specifically by two killer cell types of the adaptive and the innate immune systems; cytotoxic T lymphocytes (CTL) and natural killer (NK) cells, respectively. Another fascinating aspect of HCMV is that like other highly developed herpesviruses, it expresses its own unique set of microRNAs. Here, we initially describe how the activity of NK cells is regulated under normal conditions and during infection. Then, we discuss what is currently known about HCMV microRNA-mediated interactions, with special emphasis on immune modulation and NK cell evasion. We further illustrate the significant modulation of cellular microRNAs during HCMV infection. Although, the full target spectrum of HCMV microRNAs is far from being completely elucidated, it can already be concluded that HCMV uses its “multitasking” microRNAs to globally affect its own life cycle, as well as important cellular and immune-related pathways.




Similar content being viewed by others
References
Kiessling R, Klein E, Wigzell H (1975) “Natural” killer cells in the mouse. I. Cytotoxic cells with specificity for mouse Moloney leukemia cells. Specificity and distribution according to genotype. Eur J Immunol 5:112–117
Kiessling R, Klein E, Pross H et al (1975) “Natural” killer cells in the mouse. II. Cytotoxic cells with specificity for mouse Moloney leukemia cells. Characteristics of the killer cell. Eur J Immunol 5:117–121
Robertson MJ, Caligiuri MA, Manley TJ et al (1990) Human natural killer cell adhesion molecules. Differential expression after activation and participation in cytolysis. J Immunol 145:3194–3201
Sivori S, Vitale M, Morelli L et al (1997) p46, a novel natural killer cell-specific surface molecule that mediates cell activation. J Exp Med 186:1129–1136
Caligiuri MA (2008) Human natural killer cells. Blood 112:461–469
Galy A, Travis M, Cen D et al (1995) Human T, B, natural killer, and dendritic cells arise from a common bone marrow progenitor cell subset. Immunity 3:459–473
Seaman WE, Gindhart TD, Greenspan JS et al (1979) Natural killer cells, bone, and the bone marrow: studies in estrogen-treated mice and in congenitally osteopetrotic (mi/mi) mice. J Immunol 122:2541–2547
Kumar V, Ben-Ezra J, Bennett M et al (1979) Natural killer cells in mice treated with 89strontium: normal target-binding cell numbers but inability to kill even after interferon administration. J Immunol 123:1832–1838
Yu J, Freud AG, Caligiuri MA (2013) Location and cellular stages of natural killer cell development. Trends Immunol 34:573–582
Beziat V, Liu LL, Malmberg JA et al (2013) NK cell responses to cytomegalovirus infection lead to stable imprints in the human KIR repertoire and involve activating KIRs. Blood 121:2678–2688
Della Chiesa M, Falco M, Muccio L et al (2013) Impact of HCMV infection on NK cell development and function after HSCT. Front Immunol 4:458
Orange JS (2013) Natural killer cell deficiency. J Allergy Clin Immunol 132:515–525, quiz 526
Siren J, Sareneva T, Pirhonen J et al (2004) Cytokine and contact-dependent activation of natural killer cells by influenza A or Sendai virus-infected macrophages. J Gen Virol 85:2357–2364
Schroder K, Hertzog PJ, Ravasi T et al (2004) Interferon-gamma: an overview of signals, mechanisms and functions. J Leukoc Biol 75:163–189
Biron CA, Nguyen KB, Pien GC et al (1999) Natural killer cells in antiviral defense: function and regulation by innate cytokines. Annu Rev Immunol 17:189–220
Gur C, Porgador A, Elboim M et al (2010) The activating receptor NKp46 is essential for the development of type 1 diabetes. Nat Immunol 11:121–128
Gur C, Enk J, Kassem SA et al (2011) Recognition and killing of human and murine pancreatic beta cells by the NK receptor NKp46. J Immunol 187:3096–3103
Gur C, Enk J, Weitman E et al (2013) The expression of the beta cell-derived autoimmune ligand for the killer receptor nkp46 is attenuated in type 2 diabetes. PLoS One 8:e74033
Deniz G, van de Veen W, Akdis M (2013) Natural killer cells in patients with allergic diseases. J Allergy Clin Immunol 132:527–535
Ghadially H, Horani A, Glasner A et al (2013) NKp46 regulates allergic responses. Eur J Immunol 43:3006–3016
Hsieh CL, Obara H, Ogura Y et al (2002) NK cells and transplantation. Transpl Immunol 9:111–114
Kroemer A, Edtinger K, Li XC (2008) The innate natural killer cells in transplant rejection and tolerance induction. Curr Opin Organ Transplant 13:339–343
Uharek L, Glass B, Gaska T et al (1993) Natural killer cells as effector cells of graft-versus-leukemia activity in a murine transplantation model. Bone Marrow Transplant 12(Suppl 3):S57–S60
Hanna J, Goldman-Wohl D, Hamani Y et al (2006) Decidual NK cells regulate key developmental processes at the human fetal-maternal interface. Nat Med 12:1065–1074
Manaster I, Mizrahi S, Goldman-Wohl D et al (2008) Endometrial NK cells are special immature cells that await pregnancy. J Immunol 181:1869–1876
Topham NJ, Hewitt EW (2009) Natural killer cell cytotoxicity: how do they pull the trigger? Immunology 128:7–15
Orange JS, Ramesh N, Remold-O’Donnell E et al (2002) Wiskott-Aldrich syndrome protein is required for NK cell cytotoxicity and colocalizes with actin to NK cell-activating immunologic synapses. Proc Natl Acad Sci U S A 99:11351–11356
Oshimi Y, Oda S, Honda Y et al (1996) Involvement of Fas ligand and Fas-mediated pathway in the cytotoxicity of human natural killer cells. J Immunol 157:2909–2915
Zamai L, Ahmad M, Bennett IM et al (1998) Natural killer (NK) cell-mediated cytotoxicity: differential use of TRAIL and Fas ligand by immature and mature primary human NK cells. J Exp Med 188:2375–2380
Spaggiari GM, Carosio R, Pende D et al (2001) NK cell-mediated lysis of autologous antigen-presenting cells is triggered by the engagement of the phosphatidylinositol 3-kinase upon ligation of the natural cytotoxicity receptors NKp30 and NKp46. Eur J Immunol 31:1656–1665
Zitvogel L, Terme M, Borg C et al (2006) Dendritic cell-NK cell cross-talk: regulation and physiopathology. Curr Top Microbiol Immunol 298:157–174
Crome SQ, Lang PA, Lang KS et al (2013) Natural killer cells regulate diverse T cell responses. Trends Immunol 34:342–349
Zingoni A, Ardolino M, Santoni A et al (2012) NKG2D and DNAM-1 activating receptors and their ligands in NK-T cell interactions: role in the NK cell-mediated negative regulation of T cell responses. Front Immunol 3:408
Moretta L, Bottino C, Pende D et al (2002) Human natural killer cells: their origin, receptors and function. Eur J Immunol 32:1205–1211
Vilches C, Parham P (2002) KIR: diverse, rapidly evolving receptors of innate and adaptive immunity. Annu Rev Immunol 20:217–251
Middleton D, Curran M, Maxwell L (2002) Natural killer cells and their receptors. Transpl Immunol 10:147–164
Rajagopalan S, Long EO (2012) KIR2DL4 (CD158d): an activation receptor for HLA-G. Front Immunol 3:258
Vitale M, Castriconi R, Parolini S et al (1999) The leukocyte Ig-like receptor (LIR)-1 for the cytomegalovirus UL18 protein displays a broad specificity for different HLA class I alleles: analysis of LIR-1 + NK cell clones. Int Immunol 11:29–35
O’Callaghan CA (2000) Molecular basis of human natural killer cell recognition of HLA-E (human leucocyte antigen-E) and its relevance to clearance of pathogen-infected and tumour cells. Clin Sci (Lond) 99:9–17
Braud VM, Allan DS, Wilson D et al (1998) TAP- and tapasin-dependent HLA-E surface expression correlates with the binding of an MHC class I leader peptide. Curr Biol 8:1–10
O’Callaghan CA, Bell JI (1998) Structure and function of the human MHC class Ib molecules HLA-E, HLA-F and HLA-G. Immunol Rev 163:129–138
Carretero M, Cantoni C, Bellon T et al (1997) The CD94 and NKG2-A C-type lectins covalently assemble to form a natural killer cell inhibitory receptor for HLA class I molecules. Eur J Immunol 27:563–567
Lazetic S, Chang C, Houchins JP et al (1996) Human natural killer cell receptors involved in MHC class I recognition are disulfide-linked heterodimers of CD94 and NKG2 subunits. J Immunol 157:4741–4745
Stanietsky N, Simic H, Arapovic J et al (2009) The interaction of TIGIT with PVR and PVRL2 inhibits human NK cell cytotoxicity. Proc Natl Acad Sci U S A 106:17858–17863
Lankry D, Rovis TL, Jonjic S et al (2013) The interaction between CD300a and phosphatidylserine inhibits tumor cell killing by NK cells. Eur J Immunol 43:2151–2161
Markel G, Wolf D, Hanna J et al (2002) Pivotal role of CEACAM1 protein in the inhibition of activated decidual lymphocyte functions. J Clin Invest 110:943–953
Pende D, Parolini S, Pessino A et al (1999) Identification and molecular characterization of NKp30, a novel triggering receptor involved in natural cytotoxicity mediated by human natural killer cells. J Exp Med 190:1505–1516
Vitale M, Bottino C, Sivori S et al (1998) NKp44, a novel triggering surface molecule specifically expressed by activated natural killer cells, is involved in non-major histocompatibility complex-restricted tumor cell lysis. J Exp Med 187:2065–2072
Porgador A (2005) Natural cytotoxicity receptors: pattern recognition and involvement of carbohydrates. Sci World J 5:151–154
Bottino C, Moretta L, Pende D et al (2004) Learning how to discriminate between friends and enemies, a lesson from Natural Killer cells. Mol Immunol 41:569–575
Carrega P, Pezzino G, Queirolo P et al (2009) Susceptibility of human melanoma cells to autologous natural killer (NK) cell killing: HLA-related effector mechanisms and role of unlicensed NK cells. PLoS One 4:e8132
Halfteck GG, Elboim M, Gur C et al (2009) Enhanced in vivo growth of lymphoma tumors in the absence of the NK-activating receptor NKp46/NCR1. J Immunol 182:2221–2230
Baychelier F, Sennepin A, Ermonval M et al (2013) Identification of a cellular ligand for the natural cytotoxicity receptor NKp44. Blood 122:2935–2942
Pogge von Strandmann E, Simhadri VR, von Tresckow B et al (2007) Human leukocyte antigen-B-associated transcript 3 is released from tumor cells and engages the NKp30 receptor on natural killer cells. Immunity 27:965–974
Brandt CS, Baratin M, Yi EC et al (2009) The B7 family member B7-H6 is a tumor cell ligand for the activating natural killer cell receptor NKp30 in humans. J Exp Med 206:1495–1503
Rosental B, Brusilovsky M, Hadad U et al (2011) Proliferating cell nuclear antigen is a novel inhibitory ligand for the natural cytotoxicity receptor NKp44. J Immunol 187:5693–5702
Ferlazzo G, Tsang ML, Moretta L et al (2002) Human dendritic cells activate resting natural killer (NK) cells and are recognized via the NKp30 receptor by activated NK cells. J Exp Med 195:343–351
Seidel E, Glasner A, Mandelboim O (2012) Virus-mediated inhibition of natural cytotoxicity receptor recognition. Cell Mol Life Sci 69:3911–3920
Jarahian M, Fiedler M, Cohnen A et al (2011) Modulation of NKp30- and NKp46-mediated natural killer cell responses by poxviral hemagglutinin. PLoS Pathog 7:e1002195
Arnon TI, Achdout H, Levi O et al (2005) Inhibition of the NKp30 activating receptor by pp 65 of human cytomegalovirus. Nat Immunol 6:515–523
Bauer S, Groh V, Wu J et al (1999) Activation of NK cells and T cells by NKG2D, a receptor for stress-inducible MICA. Science 285:727–729
Houchins JP, Yabe T, McSherry C et al (1991) DNA sequence analysis of NKG2, a family of related cDNA clones encoding type II integral membrane proteins on human natural killer cells. J Exp Med 173:1017–1020
Wu J, Song Y, Bakker AB et al (1999) An activating immunoreceptor complex formed by NKG2D and DAP10. Science 285:730–732
Karimi M, Cao TM, Baker JA et al (2005) Silencing human NKG2D, DAP10, and DAP12 reduces cytotoxicity of activated CD8+ T cells and NK cells. J Immunol 175:7819–7828
Raulet DH, Gasser S, Gowen BG et al (2013) Regulation of ligands for the NKG2D activating receptor. Annu Rev Immunol 31:413–441
Groh V, Rhinehart R, Secrist H et al (1999) Broad tumor-associated expression and recognition by tumor-derived gamma delta T cells of MICA and MICB. Proc Natl Acad Sci U S A 96:6879–6884
Lakshmikanth T, Burke S, Ali TH et al (2009) NCRs and DNAM-1 mediate NK cell recognition and lysis of human and mouse melanoma cell lines in vitro and in vivo. J Clin Invest 119:1251–1263
Lanier LL (1998) NK cell receptors. Annu Rev Immunol 16:359–393
Falco M, Marcenaro E, Romeo E et al (2004) Homophilic interaction of NTBA, a member of the CD2 molecular family: induction of cytotoxicity and cytokine release in human NK cells. Eur J Immunol 34:1663–1672
Vitale M, Falco M, Castriconi R et al (2001) Identification of NKp80, a novel triggering molecule expressed by human NK cells. Eur J Immunol 31:233–242
Mathew SO, Rao KK, Kim JR et al (2009) Functional role of human NK cell receptor 2B4 (CD244) isoforms. Eur J Immunol 39:1632–1641
Fuchs A, Cella M, Giurisato E et al (2004) Cutting edge: CD96 (tactile) promotes NK cell-target cell adhesion by interacting with the poliovirus receptor (CD155). J Immunol 172:3994–3998
Orange JS (2002) Human natural killer cell deficiencies and susceptibility to infection. Microbes Infect 4:1545–1558
Martin MP, Gao X, Lee JH et al (2002) Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat Genet 31:429–434
Martin MP, Qi Y, Gao X et al (2007) Innate partnership of HLA-B and KIR3DL1 subtypes against HIV-1. Nat Genet 39:733–740
Collins KL, Chen BK, Kalams SA et al (1998) HIV-1 Nef protein protects infected primary cells against killing by cytotoxic T lymphocytes. Nature 391:397–401
Cohen GB, Gandhi RT, Davis DM et al (1999) The selective downregulation of class I major histocompatibility complex proteins by HIV-1 protects HIV-infected cells from NK cells. Immunity 10:661–671
Kiepiela P, Leslie AJ, Honeyborne I et al (2004) Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature 432:769–775
Arnon TI, Lev M, Katz G et al (2001) Recognition of viral hemagglutinins by NKp44 but not by NKp30. Eur J Immunol 31:2680–2689
Mandelboim O, Lieberman N, Lev M et al (2001) Recognition of haemagglutinins on virus-infected cells by NKp46 activates lysis by human NK cells. Nature 409:1055–1060
Gazit R, Gruda R, Elboim M et al (2006) Lethal influenza infection in the absence of the natural killer cell receptor gene Ncr1. Nat Immunol 7:517–523
Glasner A, Zurunic A, Meningher T et al (2012) Elucidating the mechanisms of influenza virus recognition by Ncr1. PLoS One 7:e36837
Moscona A (2005) Neuraminidase Inhibitors for Influenza. N Engl J Med 353:1363–1373
Tamura D, Sugaya N, Ozawa M et al (2011) Frequency of drug-resistant viruses and virus shedding in pediatric influenza patients treated with neuraminidase inhibitors. Clin Infect Dis 52:432–437
Bar-On Y, Glasner A, Meningher T et al (2013) Neuraminidase-mediated, NKp46-dependent immune-evasion mechanism of influenza viruses. Cell Rep 3:1044–1050
Bar-On Y, Seidel E, Tsukerman P et al (2014) Influenza virus uses its neuraminidase protein to evade the recognition of two activating NK cell receptors. J Infect Dis. doi:10.1093/infdis/jiu094
Ebell MH (2004) Epstein-Barr virus infectious mononucleosis. Am Fam Physician 70:1279–1287
Kutok JL, Wang F (2006) Spectrum of Epstein-Barr virus-associated diseases. Annu Rev Pathol 1:375–404
Parolini S, Bottino C, Falco M et al (2000) X-linked lymphoproliferative disease. 2B4 molecules displaying inhibitory rather than activating function are responsible for the inability of natural killer cells to kill Epstein-Barr virus-infected cells. J Exp Med 192:337–346
Shaw RK, Issekutz AC, Fraser R et al (2012) Bilateral adrenal EBV-associated smooth muscle tumors in a child with a natural killer cell deficiency. Blood 119:4009–4012
Nachmani D, Stern-Ginossar N, Sarid R et al (2009) Diverse herpesvirus microRNAs target the stress-induced immune ligand MICB to escape recognition by natural killer cells. Cell Host Microbe 5:376–385
Griffin BD, Gram AM, Mulder A et al (2013) EBV BILF1 evolved to downregulate cell surface display of a wide range of HLA class I molecules through their cytoplasmic tail. J Immunol 190:1672–1684
Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297
Borchert GM, Lanier W, Davidson BL (2006) RNA polymerase III transcribes human microRNAs. Nat Struct Mol Biol 13:1097–1101
Lee Y, Kim M, Han J et al (2004) MicroRNA genes are transcribed by RNA polymerase II. EMBO J 23:4051–4060
Lee Y, Ahn C, Han J et al (2003) The nuclear RNase III Drosha initiates microRNA processing. Nature 425:415–419
Havens MA, Reich AA, Duelli DM et al (2012) Biogenesis of mammalian microRNAs by a non-canonical processing pathway. Nucleic Acids Res 40:4626–4640
Wu Q, Song R, Ortogero N et al (2012) The RNase III enzyme DROSHA is essential for microRNA production and spermatogenesis. J Biol Chem 287:25173–25190
Landthaler M, Yalcin A, Tuschl T (2004) The human DiGeorge syndrome critical region gene 8 and its D. melanogaster homolog are required for miRNA biogenesis. Curr Biol 14:2162–2167
Gwizdek C, Ossareh-Nazari B, Brownawell AM et al (2004) Minihelix-containing RNAs mediate exportin-5-dependent nuclear export of the double-stranded RNA-binding protein ILF3. J Biol Chem 279:884–891
Redfern AD, Colley SM, Beveridge DJ et al (2013) RNA-induced silencing complex (RISC) Proteins PACT, TRBP, and Dicer are SRA binding nuclear receptor coregulators. Proc Natl Acad Sci U S A 110:6536–6541
Winter J, Jung S, Keller S et al (2009) Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol 11:228–234
Ma JB, Ye K, Patel DJ (2004) Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nature 429:318–322
Khvorova A, Reynolds A, Jayasena SD (2003) Functional siRNAs and miRNAs exhibit strand bias. Cell 115:209–216
Orom UA, Nielsen FC, Lund AH (2008) MicroRNA-10a binds the 5′UTR of ribosomal protein mRNAs and enhances their translation. Mol Cell 30:460–471
Bartel DP (2009) MicroRNAs: target recognition and regulatory functions. Cell 136:215–233
Duursma AM, Kedde M, Schrier M et al (2008) miR-148 targets human DNMT3b protein coding region. RNA 14:872–877
Stern-Ginossar N, Gur C, Biton M et al (2008) Human microRNAs regulate stress-induced immune responses mediated by the receptor NKG2D. Nat Immunol 9:1065–1073
Davison AJ, Eberle R, Ehlers B et al (2009) The order Herpesvirales. Arch Virol 154:171–177
Weller TH (1970) Review. Cytomegaloviruses: the difficult years. J Infect Dis 122:532–539
Ho M (2008) The history of cytomegalovirus and its diseases. Med Microbiol Immunol 197:65–73
Brooks GF (2013) Herpesviruses. In: Carroll KC (ed) Jawetz, Melnick, and Adelberg’s medical microbiology, McGraw-Hill, New York, p. 26e
Engman ML, Malm G, Engstrom L et al (2008) Congenital CMV infection: prevalence in newborns and the impact on hearing deficit. Scand J Infect Dis 40:935–942
Scheurer ME, Bondy ML, Aldape KD et al (2008) Detection of human cytomegalovirus in different histological types of gliomas. Acta Neuropathol 116:79–86
Bravender T (2010) Epstein-Barr virus, cytomegalovirus, and infectious mononucleosis. Adolesc Med State Art Rev 21:251–264, ix
Mamun Al M, Rahman S, Khan M (2009) Acute cytomegalovirus hepatitis in immunocompetent host. Kathmandu Univ Med J (KUMJ) 7:79–81
Azad AK, Ahmed T, Chowdhury AJ et al (2008) Cytomegalovirus induced hepatitis in an immunocompetent host. Mymensingh Med J 17:S104–S106
Emery VC (2001) Investigation of CMV disease in immunocompromised patients. J Clin Pathol 54:84–88
Cantoni N, Hirsch HH, Khanna N et al (2010) Evidence for a bidirectional relationship between cytomegalovirus replication and acute graft-versus-host disease. Biol Blood Marrow Transplant 16:1309–1314
Weekes MP, Tan SY, Poole E et al (2013) Latency-associated degradation of the MRP1 drug transporter during latent human cytomegalovirus infection. Science 340:199–202
Compton T, Feire A (2007) Early events in human cytomegalovirus infection. In: Arvin A, Campadelli-Fiume G, Mocarski E, Moore PS, Roizman B, Whitley R, Yamanishi K (eds) Human herpesviruses: biology, therapy, and immunoprophylaxis. Cambridge University Press, Cambridge
Dolan A, Cunningham C, Hector RD et al (2004) Genetic content of wild-type human cytomegalovirus. J Gen Virol 85:1301–1312
Pignatelli S, Dal Monte P, Rossini G et al (2004) Genetic polymorphisms among human cytomegalovirus (HCMV) wild-type strains. Rev Med Virol 14:383–410
Cha TA, Tom E, Kemble GW et al (1996) Human cytomegalovirus clinical isolates carry at least 19 genes not found in laboratory strains. J Virol 70:78–83
Kalejta RF (2008) Tegument proteins of human cytomegalovirus. Microbiol Mol Biol Rev 72:249–265, table of contents
Isaacson MK, Compton T (2009) Human cytomegalovirus glycoprotein B is required for virus entry and cell-to-cell spread but not for virion attachment, assembly, or egress. J Virol 83:3891–3903
Pietropaolo RL, Compton T (1997) Direct interaction between human cytomegalovirus glycoprotein B and cellular annexin II. J Virol 71:9803–9807
Pietropaolo R, Compton T (1999) Interference with annexin II has no effect on entry of human cytomegalovirus into fibroblast cells. J Gen Virol 80(Pt 7):1807–1816
Wille PT, Wisner TW, Ryckman B et al (2013) Human cytomegalovirus (HCMV) glycoprotein gB promotes virus entry in trans acting as the viral fusion protein rather than as a receptor-binding protein. MBio 4:e00332–00313
Ryckman BJ, Jarvis MA, Drummond DD et al (2006) Human cytomegalovirus entry into epithelial and endothelial cells depends on genes UL128 to UL150 and occurs by endocytosis and low-pH fusion. J Virol 80:710–722
Hahn G, Jores R, Mocarski ES (1998) Cytomegalovirus remains latent in a common precursor of dendritic and myeloid cells. Proc Natl Acad Sci U S A 95:3937–3942
Taylor-Wiedeman J, Sissons JG, Borysiewicz LK et al (1991) Monocytes are a major site of persistence of human cytomegalovirus in peripheral blood mononuclear cells. J Gen Virol 72(Pt 9):2059–2064
Reeves MB, MacAry PA, Lehner PJ et al (2005) Latency, chromatin remodeling, and reactivation of human cytomegalovirus in the dendritic cells of healthy carriers. Proc Natl Acad Sci U S A 102:4140–4145
Sinclair J (2010) Chromatin structure regulates human cytomegalovirus gene expression during latency, reactivation and lytic infection. Biochim Biophys Acta 1799:286–295
Davison AJ, Dolan A, Akter P et al (2003) The human cytomegalovirus genome revisited: comparison with the chimpanzee cytomegalovirus genome. J Gen Virol 84:17–28
Murphy E, Rigoutsos I, Shibuya T et al (2003) Reevaluation of human cytomegalovirus coding potential. Proc Natl Acad Sci U S A 100:13585–13590
Stern-Ginossar N, Weisburd B, Michalski A et al (2012) Decoding human cytomegalovirus. Science 338:1088–1093
Rolle A, Mousavi-Jazi M, Eriksson M et al (2003) Effects of human cytomegalovirus infection on ligands for the activating NKG2D receptor of NK cells: up-regulation of UL16-binding protein (ULBP)1 and ULBP2 is counteracted by the viral UL16 protein. J Immunol 171:902–908
Eagle RA, Traherne JA, Hair JR et al (2009) ULBP6/RAET1L is an additional human NKG2D ligand. Eur J Immunol 39:3207–3216
Wu J, Chalupny NJ, Manley TJ et al (2003) Intracellular retention of the MHC class I-related chain B ligand of NKG2D by the human cytomegalovirus UL16 glycoprotein. J Immunol 170:4196–4200
Ashiru O, Bennett NJ, Boyle LH et al (2009) NKG2D ligand MICA is retained in the cis-Golgi apparatus by human cytomegalovirus protein UL142. J Virol 83:12345–12354
Fielding CA, Aicheler R, Stanton RJ et al (2014) Two novel human cytomegalovirus NK cell evasion functions target MICA for lysosomal degradation. PLoS Pathog 10:e1004058
Bennett NJ, Ashiru O, Morgan FJ et al (2010) Intracellular sequestration of the NKG2D ligand ULBP3 by human cytomegalovirus. J Immunol 185:1093–1102
Hewitt EW, Gupta SS, Lehner PJ (2001) The human cytomegalovirus gene product US6 inhibits ATP binding by TAP. EMBO J 20:387–396
van der Wal FJ, Kikkert M, Wiertz E (2002) The HCMV gene products US2 and US11 target MHC class I molecules for degradation in the cytosol. Curr Top Microbiol Immunol 269:37–55
Noriega VM, Hesse J, Gardner TJ et al (2012) Human cytomegalovirus US3 modulates destruction of MHC class I molecules. Mol Immunol 51:245–253
Furman MH, Dey N, Tortorella D et al (2002) The human cytomegalovirus US10 gene product delays trafficking of major histocompatibility complex class I molecules. J Virol 76:11753–11756
Trgovcich J, Cebulla C, Zimmerman P et al (2006) Human cytomegalovirus protein pp 71 disrupts major histocompatibility complex class I cell surface expression. J Virol 80:951–963
Chapman TL, Heikeman AP, Bjorkman PJ (1999) The inhibitory receptor LIR-1 uses a common binding interaction to recognize class I MHC molecules and the viral homolog UL18. Immunity 11:603–613
Pfeffer S, Zavolan M, Grasser FA et al (2004) Identification of virus-encoded microRNAs. Science 304:734–736
Pfeffer S, Sewer A, Lagos-Quintana M et al (2005) Identification of microRNAs of the herpesvirus family. Nat Methods 2:269–276
Cui C, Griffiths A, Li G et al (2006) Prediction and identification of herpes simplex virus 1-encoded microRNAs. J Virol 80:5499–5508
Sun L, Li Q (2012) The miRNAs of Herpes Simplex Virus (HSV). Virol Sin 27:332–337
Grundhoff A, Sullivan CS (2011) Virus-encoded microRNAs. Virology 411:325–343
Kozomara A, Griffiths-Jones S (2014) miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res 42:D68–D73
Stern-Ginossar N, Elefant N, Zimmermann A et al (2007) Host immune system gene targeting by a viral miRNA. Science 317:376–381
Bauman Y, Nachmani D, Vitenshtein A et al (2011) An identical miRNA of the human JC and BK polyoma viruses targets the stress-induced ligand ULBP3 to escape immune elimination. Cell Host Microbe 9:93–102
Kim S, Lee S, Shin J et al (2011) Human cytomegalovirus microRNA miR-US4-1 inhibits CD8(+) T cell responses by targeting the aminopeptidase ERAP1. Nat Immunol 12:984–991
Tomasec P, Braud VM, Rickards C et al (2000) Surface expression of HLA-E, an inhibitor of natural killer cells, enhanced by human cytomegalovirus gpUL40. Science 287:1031
Nachmani D, Zimmermann A, Oiknine Djian E et al (2014) MicroRNA editing facilitates immune elimination of HCMV infected cells. PLoS Pathog 10:e1003963
Maghazachi AA, al-Aoukaty A, Schall TJ (1994) C-C chemokines induce the chemotaxis of NK and IL-2-activated NK cells. Role for G proteins. J Immunol 153:4969–4977
Loetscher P, Seitz M, Clark-Lewis I et al (1996) Activation of NK cells by CC chemokines. Chemotaxis, Ca2+ mobilization, and enzyme release. J Immunol 156:322–327
Taylor RT, Bresnahan WA (2006) Human cytomegalovirus immediate-early 2 protein IE86 blocks virus-induced chemokine expression. J Virol 80:920–928
Bodaghi B, Jones TR, Zipeto D et al (1998) Chemokine sequestration by viral chemoreceptors as a novel viral escape strategy: withdrawal of chemokines from the environment of cytomegalovirus-infected cells. J Exp Med 188:855–866
Wang D, Bresnahan W, Shenk T (2004) Human cytomegalovirus encodes a highly specific RANTES decoy receptor. Proc Natl Acad Sci U S A 101:16642–16647
Kim Y, Lee S, Kim S et al (2012) Human cytomegalovirus clinical strain-specific microRNA miR-UL148D targets the human chemokine RANTES during infection. PLoS Pathog 8:e1002577
Sarras H, Alizadeh Azami S, McPherson JP (2010) In search of a function for BCLAF1. Sci World J 10:1450–1461
Lee YY, Yu YB, Gunawardena HP et al (2012) BCLAF1 is a radiation-induced H2AX-interacting partner involved in gammaH2AX-mediated regulation of apoptosis and DNA repair. Cell Death Dis 3:e359
Lee SH, Kalejta RF, Kerry J et al (2012) BclAF1 restriction factor is neutralized by proteasomal degradation and microRNA repression during human cytomegalovirus infection. Proc Natl Acad Sci U S A 109:9575–9580
Ziegelbauer JM, Sullivan CS, Ganem D (2009) Tandem array-based expression screens identify host mRNA targets of virus-encoded microRNAs. Nat Genet 41:130–134
Park MH, Song MJ, Cho MC et al (2012) Interleukin-32 enhances cytotoxic effect of natural killer cells to cancer cells via activation of death receptor 3. Immunology 135:63–72
Cheon S, Lee JH, Park S et al (2011) Overexpression of IL-32alpha increases natural killer cell-mediated killing through up-regulation of Fas and UL16-binding protein 2 (ULBP2) expression in human chronic myeloid leukemia cells. J Biol Chem 286:12049–12055
Huang Y, Qi Y, Ma Y et al (2013) The expression of interleukin-32 is activated by human cytomegalovirus infection and down regulated by hcmv-miR-UL112-1. Virol J 10:51
Hook LM, Grey F, Grabski R et al (2014) Cytomegalovirus miRNAs target secretory pathway genes to facilitate formation of the virion assembly compartment and reduce cytokine secretion. Cell Host Microbe 15:363–373
Pavelin J, Reynolds N, Chiweshe S et al (2013) Systematic microRNA analysis identifies ATP6V0C as an essential host factor for human cytomegalovirus replication. PLoS Pathog 9:e1003820
Grey F, Tirabassi R, Meyers H et al (2010) A viral microRNA down-regulates multiple cell cycle genes through mRNA 5′UTRs. PLoS Pathog 6:e1000967
Qi M, Qi Y, Ma Y et al (2013) Over-expression of human cytomegalovirus miR-US25-2-3p downregulates eIF4A1 and inhibits HCMV replication. FEBS Lett 587:2266–2271
Stern-Ginossar N, Saleh N, Goldberg MD et al (2009) Analysis of human cytomegalovirus-encoded microRNA activity during infection. J Virol 83:10684–10693
Grey F, Meyers H, White EA et al (2007) A human cytomegalovirus-encoded microRNA regulates expression of multiple viral genes involved in replication. PLoS Pathog 3:e163
Li S, Zhu J, Zhang W et al (2011) Signature microRNA expression profile of essential hypertension and its novel link to human cytomegalovirus infection. Circulation 124:175–184
Yamamoto T, Suzuki S, Radsak K et al (1998) The UL112/113 gene products of human cytomegalovirus which colocalize with viral DNA in infected cell nuclei are related to efficient viral DNA replication. Virus Res 56:107–114
Wells R, Stensland L, Vieira J (2009) The human cytomegalovirus UL112-113 locus can activate the full Kaposi’s sarcoma-associated herpesvirus lytic replication cycle. J Virol 83:4695–4699
Lee S, Song J, Kim S et al (2013) Selective degradation of host MicroRNAs by an intergenic HCMV noncoding RNA accelerates virus production. Cell Host Microbe 13:678–690
Wang FZ, Weber F, Croce C et al (2008) Human cytomegalovirus infection alters the expression of cellular microRNA species that affect its replication. J Virol 82:9065–9074
Poole E, McGregor Dallas SR, Colston J et al (2011) Virally induced changes in cellular microRNAs maintain latency of human cytomegalovirus in CD34(+) progenitors. J Gen Virol 92:1539–1549
Sinclair JH, Reeves MB (2013) Human cytomegalovirus manipulation of latently infected cells. Viruses 5:2803–2824
Huang HC, Yu HR, Huang LT et al (2012) miRNA-125b regulates TNF-alpha production in CD14+ neonatal monocytes via post-transcriptional regulation. J Leukoc Biol 92:171–182
Santhakumar D, Forster T, Laqtom NN et al (2010) Combined agonist–antagonist genome-wide functional screening identifies broadly active antiviral microRNAs. Proc Natl Acad Sci U S A 107:13830–13835
Zhang S, Liu L, Wang R et al (2013) MiR-199a-5p promotes migration and tube formation of human cytomegalovirus-infected endothelial cells through downregulation of SIRT1 and eNOS. Arch Virol 158:2443–2452
Zhang S, Liu L, Wang R et al (2013) MicroRNA-217 promotes angiogenesis of human cytomegalovirus-infected endothelial cells through downregulation of SIRT1 and FOXO3A. PLoS One 8:e83620
Menghini R, Casagrande V, Cardellini M et al (2009) MicroRNA 217 modulates endothelial cell senescence via silent information regulator 1. Circulation 120:1524–1532
Pecot CV, Rupaimoole R, Yang D et al (2013) Tumour angiogenesis regulation by the miR-200 family. Nat Commun 4:2427
Haneklaus M, Gerlic M, O’Neill LA et al (2013) miR-223: infection, inflammation and cancer. J Intern Med 274:215–226
Fu M, Gao Y, Zhou Q et al (2014) Human cytomegalovirus latent infection alters the expression of cellular and viral microRNA. Gene 536:272–278
Stark TJ, Arnold JD, Spector DH et al (2012) High-resolution profiling and analysis of viral and host small RNAs during human cytomegalovirus infection. J Virol 86:226–235
Xiong Y, Fang JH, Yun JP et al (2010) Effects of microRNA-29 on apoptosis, tumorigenicity, and prognosis of hepatocellular carcinoma. Hepatology 51:836–845
Mott JL, Kobayashi S, Bronk SF et al (2007) mir-29 regulates Mcl-1 protein expression and apoptosis. Oncogene 26:6133–6140
Li Y, Wang H, Tao K et al (2013) miR-29b suppresses CML cell proliferation and induces apoptosis via regulation of BCR/ABL1 protein. Exp Cell Res 319:1094–1101
O’Connor CM, Vanicek J, Murphy EA (2014) Host MicroRNA regulation of human cytomegalovirus immediate early protein translation promotes viral latency. J Virol 88:5524–5532
Dolken L, Pfeffer S, Koszinowski UH (2009) Cytomegalovirus microRNAs. Virus Genes 38:355–364
Acknowledgments
This study was supported by funding from the European Research Council under the European Union’s Seventh Framework Program (FP/2007-2013)/ERC Grant Agreement number 320473-BacNK. Further support came from the GIF foundation, from the Lewis family foundation, the ICRF professorship grant, the Israeli Science Foundation, the Helmhotz Association, and the Rosetrees Trust (all to O.M.). O.M. is a Crown Professor of Molecular Immunology.
Author information
Authors and Affiliations
Corresponding author
Additional information
This article is a contribution to the special issue on Immune Modulation, properties and models of CMV - Guest Editor: Ofer Mandelboim
Rights and permissions
About this article
Cite this article
Goldberger, T., Mandelboim, O. The use of microRNA by human viruses: lessons from NK cells and HCMV infection. Semin Immunopathol 36, 659–674 (2014). https://doi.org/10.1007/s00281-014-0447-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00281-014-0447-3