Abstract
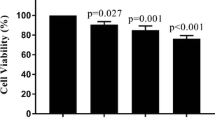
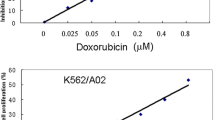
Dexamethasone is considered as a direct chemotherapeutic agent in the treatment of pediatric acute lymphoblastic leukemia (ALL). Beside the advantages of the drug, some problems arising from the dose-related side effects are challenging issues during the treatment. Accordingly, the classification of patients to dexamethasone sensitive and resistance groups can help to select optimizing the therapeutic dose with the lowest adverse effects particularly in sensitive cases. For this purpose, we investigated inhibited proliferation and induced cytotoxicity in NALM-6 cells, as sensitive cells, after dexamethasone treatment. In addition, comparative protein expression analysis using the 2DE–MALDI–TOF MS technique was performed to identify the specific altered proteins. In addition, we evaluated mRNA expression levels of the identified proteins in bone-marrow samples from pediatric ALL patients using the real-time q-PCR method. Eventually, proteomic analysis revealed a combination of biomarkers, including capping proteins (CAPZA1 and CAPZB), chloride channel (CLIC1), purine nucleoside phosphorylase (PNP), and proteasome activator (PSME1), in response to the dexamethasone treatment. In addition, our results indicated low expression of identified proteins at both the mRNA and protein expression levels after drug treatment. Moreover, quantitative real-time PCR data analysis indicated that independent of the molecular subtypes of the leukemia, CAPZA1, CAPZB, CLIC1, and PNP expression levels were lower in ALL samples than normal samples, although PSME1 expression level was higher in ALL samples than normal samples. Furthermore, the expression level of all proteins (except PSME1) was different between high-risk and standard-risk patients that suggesting the prognostic value of them. In conclusion, our study suggests a panel of biomarkers comprising CAPZA1, CAPZB, CLIC1, PNP, and PSME1 as early diagnosis and treatment evaluation markers that may differentiate cancer cells which are presumably to benefit from dexamethasone-based chemotherapy and may facilitate the prediction of clinical outcome.





Similar content being viewed by others
References
Inaba H, Pui CH (2010) Glucocorticoid use in acute lymphoblastic leukaemia. Lancet Oncol 11(11):1096–1106
Harmon JM, Norman MR, Fowlkes BJ, Thompson EB (1979) Dexamethasone induces irreversible G1 arrest and death of a human lymphoid cell line. J Cell Physiol 98(2):267–278
Strauss AJ, Su JT, Dalton VM, Gelber RD, Sallan SE, Silverman LB (2001) Bony morbidity in children treated for acute lymphoblastic leukemia. J Clin Oncol 19:3066–3072
Gaynon PS, Angiolillo AL, Carroll WL et al (2010) Long-term results of the children’s cancer group studies for childhood acute lymphoblastic leukemia 1983-2002: a Children’s Oncology Group Report. Leukemia 24(2):285–297
Pui C-H, Mullighan CG, Evans WE, Relling MV (2012) Pediatric acute lymphoblastic leukemia: where are we going and how do we get there? Blood 120(6):1165–1174
Cooper SL, Brown PA (2015) Treatment of pediatric acute lymphoblastic leukemia. Pediatr Clin N Am 62(1):61–73
Kalia M (2015) Biomarkers for personalized oncology: recent advances and future challenges. Metabolism 64(3 Suppl 1):S16–S21
Minasian L, Rosen O, Auclair D, Rahman A, Pazdur R, Schilsky RL (2014) Optimizing dosing of oncology drugs. Clin Pharmacol Ther 96:572–579
Schwaederle M, Zhao M, Lee JJ (2015) Impact of precision medicine in diverse cancers: a meta-analysis of phase II clinical trials. J Clin Oncol 33(32):3817–3825
López Villar E, Wu D, Cho WC, Madero L, Wang X (2014) Proteomics-based discovery of biomarkers for paediatric acute lymphoblastic leukaemia: challenges and opportunities. J Cell Mol Med 18(7):1239–1246
Dehghan-Nayeri N, Rezaei-Tavirani M, Omrani MD, Gharehbaghian A, Goudarzi Pour K, Eshghi P (2016) Identification of potential predictive markers of dexamethasone resistance in childhood acute lymphoblastic leukemia. J Cell Commun Signal. doi:10.1007/s12079-016-0357-3
Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:e45
Mi N, Chen Y, Wang S et al (2015) CapZ regulates autophagosomal membrane shaping by promoting actin assembly inside the isolation membrane. Nat Cell Biol 17(9):1112–1123
Sun D, Zhou M, Kowolik CM et al (2011) Differential expression patterns of capping protein, protein phosphatase 1, and casein kinase 1 may serve as diagnostic markers for malignant melanoma. Melanoma Res 21(4):335–343
Cooper JA, Sept D (2008) New insights into mechanism and regulation of actin capping protein. Int Rev Cell Mol Biol 267:183–206
Feng S, Lin S, Zou J, Wang Y, Ji Q, Lv Z (2015) Association between rs12045440 polymorphism in the CAPZB intron and serum TSH concentrations in Chinese thyroid tumor patients. Int J Endocrinol 2015. doi:10.1155/2015/250542
Lee Y-J, Jeong S-H, Hong S-C et al (2013) Prognostic value of CAPZA1 overexpression in gastric cancer. Int J Oncol 42(5):1569–1577
Thompson CC, Ashcroft FJ, Patel S et al (2007) Pancreatic cancer cells overexpress gelsolin family-capping proteins, which contribute to their cell motility. Gut 56(1):95–106
Wang L, He S, Tu Y et al (2012) Elevated expression of chloride intracellular channel 1 is correlated with poor prognosis in human gliomas. J Exp Clin Cancer Res 31:44
Tung JJ, Kitajewski J (2010) Chloride intracellular channel 1 functions in endothelial cell growth and migration. J Angiogenes Res 2:23
Peretti M, Angelini M, Savalli N, Florio T, Yuspa SH, Mazzanti M (2015) Chloride channels in cancer: focus on chloride intracellular channel 1 and 4 (CLIC1 AND CLIC4) proteins in tumor development and as novel therapeutic targets. Biochim Biophys Acta 1848(10 Pt B):2523–2531
Tian Y, Guan Y, Jia Y, Meng Q, Yang J (2014) Chloride intracellular channel 1 regulates prostate cancer cell proliferation and migration through the MAPK/ERK pathway. Cancer Biother Radiopharm 29(8):339–344
Ding Q, Li M, Wu X et al (2015) CLIC1 overexpression is associated with poor prognosis in gallbladder cancer. Tumour Biol 36(1):193–198
Wang P, Zhang C, Yu P et al (2012) Regulation of colon cancer cell migration and invasion by CLIC1-mediated RVD. Mol Cell Biochem 365(1–2):313–321
Ma PF, Chen JQ, Wang Z, Liu JL, Li BP (2012) Function of chloride intracellular channel 1 in gastric cancer cells. World J Gastroenterol 18(24):3070–3080
Wang W, Xu X, Wang W et al (2011) The expression and clinical significance of CLIC1 and HSP27 in lung adenocarcinoma. Tumour Biol 32(10):1199–1208
Giuliani P, Zuccarini M, Buccella S et al (2016) Development of a new HPLC method using fluorescence detection without derivatization for determining purine nucleoside phosphorylase activity in human plasma. J Chromatogr B 15(1009–1010):114–121
Balakrishnan K, Verma D, O’Brien S et al (2010) Phase 2 and pharmacodynamic study of oral forodesine in patients with advanced, fludarabine-treated chronic lymphocytic leukemia. Blood 116:886–892
Kojima S, Chiyomaru T, Kawakami K et al (2012) Tumour suppressors miR-1 and miR-133a target the oncogenic function of purine nucleoside phosphorylase (PNP) in prostate cancer. Br J Cancer 106(2):405–413
Yamasaki T, Yoshino H, Enokida H et al (2012) Novel molecular targets regulated by tumor suppressors microRNA-1 and microRNA-133a in bladder cancer. Int J Oncol 40(6):1821–1830
Sanfilippo O, Camici M, Tozzi MG et al (1994) Relationship between the levels of purine salvage pathway enzymes and clinical/biological aggressiveness of human colon carcinoma. Cancer Biochem Biophys 14(1):57–66
Pavlou MP, Dimitromanolakis A, Martinez-Morillo E, Smid M, Foekens JA, Diamandis EP (2014) Integrating meta-analysis of microarray data and targeted proteomics for biomarker identification: application in breast cancer. J Proteome Res 13(6):2897–2909
Zhang L, Hou Y, Wu K, Li D (2012) Comparative proteomics analysis of chronic atrophic gastritis: changes of protein expression in chronic atrophic gastritis without Helicobacter pylori infection. Braz J Med Biol Res 45(3):273–283
Zhang D, Lim SG, Koay ES (2007) Proteomic identification of down-regulation of oncoprotein DJ-1 and proteasome activator subunit 1 in hepatitis B virus-infected well-differentiated hepatocellular carcinoma. Int J Oncol 31:577–584
Miyagi T, Tatsumi T, Takehara T et al (2003) Impaired expression of proteasome subunits and human leukocyte antigens class I in human colon cancer cells. J Gastroenterol Hepatol 18:32–40
Lemaire R, Menguellet SA, Stauber J et al (2007) Specific MALDI imaging and profiling for biomarker hunting and validation: fragment of the 11S proteasome activator complex, Reg alpha fragment is a new potential ovary cancer biomarker. J Proteome Res 6:4127–4134
Zhang J, Wang K, Zhang J, Liu SS, Dai L, Zhang JY (2011) Using proteomic approach to identify tumor-associated proteins as biomarkers in human esophageal squamous cell carcinoma. J Proteome Res 10(6):2863–2872
Sánchez-Martín D, Martínez-Torrecuadrada J, Teesalu T et al (2013) Proteasome activator complex PA28 identified as an accessible target in prostate cancer by in vivo selection of human antibodies. Proc Natl Acad Sci USA 110(34):13791–13796
Muñiz Lino MA, Palacios-Rodríguez Y, Rodríguez-Cuevas S et al (2014) Comparative proteomic profiling of triple-negative breast cancer reveals that up-regulation of RhoGDI-2 is associated to the inhibition of caspase 3 and caspase 9. J Proteomics 5(111):198–211
Acknowledgements
This work was supported by the Shahid Beheshti University of Medical Sciences, Tehran, Iran.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical standards
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Rights and permissions
About this article
Cite this article
Dehghan-Nayeri, N., Eshghi, P., Pour, K.G. et al. Differential expression pattern of protein markers for predicting chemosensitivity of dexamethasone-based chemotherapy of B cell acute lymphoblastic leukemia. Cancer Chemother Pharmacol 80, 177–185 (2017). https://doi.org/10.1007/s00280-017-3347-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00280-017-3347-0