Abstract
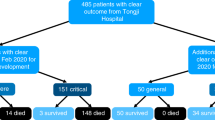
Infection post-hematopoietic stem cell transplantation (HSCT) is one of the main causes of patient mortality. Fever is the most crucial clinical symptom indicating infection. However, current microbial detection methods are limited. Therefore, timely diagnosis of infectious fever and administration of antimicrobial drugs can effectively reduce patient mortality. In this study, serum samples were collected from 181 patients with HSCT with or without infection, as well as the clinical information. And more than 80 infectious-related microRNAs in the serum were selected according to the bulk RNA-seq result and detected in the 345 time-pointed serum samples by Q-PCR. Unsupervised clustering result indicates a close association between these microRNAs expression and infection occurrence. Compared to the uninfected cohort, more than 10 serum microRNAs were identified as the combined diagnostic markers in one formula constructed by the Random Forest (RF) algorithms, with a diagnostic accuracy more than 0.90. Furthermore, correlations of serum microRNAs to immune cells, inflammatory factors, pathgens, infection tissue, and prognosis were analyzed in the infection cohort. Overall, this study demonstrates that the combination of serum microRNAs detection and machine learning algorithms holds promising potential in diagnosing infectious fever after HSCT.





Similar content being viewed by others
Data availability
The raw Illumina read data, qPCR raw data, and the group information data for all samples, as well as the bioinformatic analysis code, can be obtained upon request from the authors.
References
Arango M, Combariza JF (2017) Fever after peripheral blood stem cell infusion in haploidentical transplantation with post-transplant cyclophosphamide. Hematol Oncol Stem Cell Ther 10(2):79–84. https://doi.org/10.1016/j.hemonc.2017.03.001
Fayard A, Daguenet E, Blaise D, Chevallier P, Labussiere H, Berceanu A, Yakoub-Agha I, Socie G, Charbonnier A, Suarez F, Huynh A, Mercier M, Bulabois CE, Lioure B, Chantepie S, Beguin Y, Bourhis JH, Malfuson JV, Clement L, de la Peffault R, Cornillon J (2019) Evaluation of infectious complications after haploidentical hematopoietic stem cell transplantation with post-transplant cyclophosphamide following reduced-intensity and myeloablative conditioning: a study on behalf of the Francophone Society of Stem Cell Transplantation and Cellular Therapy (SFGM-TC). Bone Marrow Transpl 54(10):1586–1594. https://doi.org/10.1038/s41409-019-0475-7
Ullmann AJ, Schmidt-Hieber M, Bertz H, Heinz WJ, Kiehl M, Kruger W, Mousset S, Neuburger S, Neumann S, Penack O, Silling G, Vehreschild JJ, Einsele H, Maschmeyer G, Infectious Diseases Working Party of the German Society for H, Medical O, the D-K (2016) Infectious diseases in allogeneic haematopoietic stem cell transplantation: prevention and prophylaxis strategy guidelines 2016. Ann Hematol 95 (9):1435–1455. https://doi.org/10.1007/s00277-016-2711-1
Sava M, Battig V, Gerull S, Passweg JR, Khanna N, Garzoni C, Gerber B, Mueller NJ, Schanz U, Berger C, Chalandon Y, van Delden C, Neofytos D, Stampf S, Franzeck FC, Weisser M, Swiss Transplant Cohort S (2023) Bloodstream infections in allogeneic haematopoietic cell recipients from the Swiss transplant cohort study: trends of causative pathogens and resistance rates. Bone Marrow Transpl 58(1):115–118. https://doi.org/10.1038/s41409-022-01851-y
Coda J, Raser K, Anand SM, Ghosh M, Gregg K, Li J, Maciejewski JJ, Pawarode A, Riwes MM, Tillman C, Polk A, Kandarpa M, Talpaz M, Choi SW, Yanik GA, Magenau JM, Pianko MJ (2023) Pneumocystis jirovecii infection in autologous hematopoietic stem cell transplant recipients. Bone Marrow Transpl 58(4):446–451. https://doi.org/10.1038/s41409-022-01906-0
Massaro KS, Macedo R, de Castro BS, Dulley F, Oliveira MS, Yasuda MA, Levin AS, Costa SF (2014) Risk factor for death in hematopoietic stem cell transplantation: are biomarkers useful to foresee the prognosis in this population of patients? Infection 42(6):1023–1032. https://doi.org/10.1007/s15010-014-0685-2
Didelot X, Walker AS, Peto TE, Crook DW, Wilson DJ (2016) Within-host evolution of bacterial pathogens. Nat Rev Microbiol 14(3):150–162. https://doi.org/10.1038/nrmicro.2015.13
Zhou X, Li X, Wu M (2018) miRNAs reshape immunity and inflammatory responses in bacterial infection. Signal Transduct Target Ther 3:14. https://doi.org/10.1038/s41392-018-0006-9
Ma C, Li Y, Li M, Deng G, Wu X, Zeng J, Hao X, Wang X, Liu J, Cho WC, Liu X, Wang Y (2014) microRNA-124 negatively regulates TLR signaling in alveolar macrophages in response to mycobacterial infection. Mol Immunol 62(1):150–158. https://doi.org/10.1016/j.molimm.2014.06.014
Ma C, Li Y, Zeng J, Wu X, Liu X, Wang Y (2014) Mycobacterium bovis BCG triggered MyD88 induces miR-124 feedback negatively regulates immune response in alveolar epithelial cells. PLoS ONE 9(4):e92419. https://doi.org/10.1371/journal.pone.0092419
Benz F, Tacke F, Luedde M, Trautwein C, Luedde T, Koch A, Roderburg C (2015) Circulating microRNA-223 serum levels do not predict sepsis or survival in patients with critical illness. Dis Markers 2015:384208. https://doi.org/10.1155/2015/384208
Jenike AE, Halushka MK (2021) miR-21: a non-specific biomarker of all maladies. Biomark Res 9(1):18. https://doi.org/10.1186/s40364-021-00272-1
Zhang J, Li S, Li L, Li M, Guo C, Yao J, Mi S (2015) Exosome and exosomal microRNA: trafficking, sorting, and function. Genomics Proteom Bioinf 13(1):17–24. https://doi.org/10.1016/j.gpb.2015.02.001
Fabbri M, Paone A, Calore F, Galli R, Gaudio E, Santhanam R, Lovat F, Fadda P, Mao C, Nuovo GJ, Zanesi N, Crawford M, Ozer GH, Wernicke D, Alder H, Caligiuri MA, Nana-Sinkam P, Perrotti D, Croce CM (2012) MicroRNAs bind to toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci U S A 109(31):E2110–2116. https://doi.org/10.1073/pnas.1209414109
Vignard V, Labbe M, Marec N, Andre-Gregoire G, Jouand N, Fonteneau JF, Labarriere N, Fradin D (2020) MicroRNAs in Tumor Exosomes Drive Immune escape in Melanoma. Cancer Immunol Res 8(2):255–267. https://doi.org/10.1158/2326-6066.CIR-19-0522
Ye SB, Li ZL, Luo DH, Huang BJ, Chen YS, Zhang XS, Cui J, Zeng YX, Li J (2014) Tumor-derived exosomes promote tumor progression and T-cell dysfunction through the regulation of enriched exosomal microRNAs in human nasopharyngeal carcinoma. Oncotarget 5(14):5439–5452. https://doi.org/10.18632/oncotarget.2118
Mittelbrunn M, Gutierrez-Vazquez C, Villarroya-Beltri C, Gonzalez S, Sanchez-Cabo F, Gonzalez MA, Bernad A, Sanchez-Madrid F (2011) Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat Commun 2:282. https://doi.org/10.1038/ncomms1285
Ogata-Kawata H, Izumiya M, Kurioka D, Honma Y, Yamada Y, Furuta K, Gunji T, Ohta H, Okamoto H, Sonoda H, Watanabe M, Nakagama H, Yokota J, Kohno T, Tsuchiya N (2014) Circulating exosomal microRNAs as biomarkers of colon cancer. PLoS ONE 9(4):e92921. https://doi.org/10.1371/journal.pone.0092921
Huang X, Yuan T, Tschannen M, Sun Z, Jacob H, Du M, Liang M, Dittmar RL, Liu Y, Liang M, Kohli M, Thibodeau SN, Boardman L, Wang L (2013) Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genomics 14:319. https://doi.org/10.1186/1471-2164-14-319
Khandelwal A, Sharma U, Barwal TS, Seam RK, Gupta M, Rana MK, Vasquez KM, Jain A (2021) Circulating miR-320a acts as a tumor suppressor and prognostic factor in non-small cell Lung Cancer. Front Oncol 11:645475. https://doi.org/10.3389/fonc.2021.645475
Dong P, Mai Y, Zhang Z, Mi L, Wu G, Chu G, Yang G, Sun S (2014) MiR-15a/b promote adipogenesis in porcine pre-adipocyte via repressing FoxO1. Acta Biochim Biophys Sin (Shanghai) 46(7):565–571. https://doi.org/10.1093/abbs/gmu043
Wendlandt EB, Graff JW, Gioannini TL, McCaffrey AP, Wilson ME (2012) The role of microRNAs miR-200b and miR-200c in TLR4 signaling and NF-kappaB activation. Innate Immun 18(6):846–855. https://doi.org/10.1177/1753425912443903
Gu Y, Liu H, Kong F, Ye J, Jia X, Zhang Z, Li N, Yin J, Zheng G, He Z (2018) miR-22/KAT6B axis is a chemotherapeutic determiner via regulation of PI3k-Akt-NF-kB pathway in tongue squamous cell carcinoma. J Exp Clin Cancer Res 37(1):164. https://doi.org/10.1186/s13046-018-0834-z
Fan K, Spassova I, Gravemeyer J, Ritter C, Horny K, Lange A, Gambichler T, Odum N, Schrama D, Schadendorf D, Ugurel S, Becker JC (2021) Merkel cell carcinoma-derived exosome-shuttle miR-375 induces fibroblast polarization by inhibition of RBPJ and p53. Oncogene 40(5):980–996. https://doi.org/10.1038/s41388-020-01576-6
Xu G, Ding Z, Shi HF (2019) The mechanism of miR-889 regulates osteogenesis in human bone marrow mesenchymal stem cells. J Orthop Surg Res 14(1):366. https://doi.org/10.1186/s13018-019-1399-z
Mu J, Zhu D, Shen Z, Ning S, Liu Y, Chen J, Li Y, Li Z (2017) The repressive effect of miR-148a on Wnt/beta-catenin signaling involved in glabridin-induced anti-angiogenesis in human breast cancer cells. BMC Cancer 17(1):307. https://doi.org/10.1186/s12885-017-3298-1
Xu E, Zhao J, Ma J, Wang C, Zhang C, Jiang H, Cheng J, Gao R, Zhou X (2016) miR-146b-5p promotes invasion and metastasis contributing to chemoresistance in osteosarcoma by targeting zinc and ring finger 3. Oncol Rep 35(1):275–283. https://doi.org/10.3892/or.2015.4393
Lee SI, Celik S, Logsdon BA, Lundberg SM, Martins TJ, Oehler VG, Estey EH, Miller CP, Chien S, Dai J, Saxena A, Blau CA, Becker PS (2018) A machine learning approach to integrate big data for precision medicine in acute myeloid leukemia. Nat Commun 9(1):42. https://doi.org/10.1038/s41467-017-02465-5
Zoabi Y, Deri-Rozov S, Shomron N (2021) Machine learning-based prediction of COVID-19 diagnosis based on symptoms. NPJ Digit Med 4(1):3. https://doi.org/10.1038/s41746-020-00372-6
Ardakani AA, Kanafi AR, Acharya UR, Khadem N, Mohammadi A (2020) Application of deep learning technique to manage COVID-19 in routine clinical practice using CT images: results of 10 convolutional neural networks. Comput Biol Med 121:103795. https://doi.org/10.1016/j.compbiomed.2020.103795
Hobby GP, Karaduta O, Dusio GF, Singh M, Zybailov BL, Arthur JM (2019) Chronic kidney disease and the gut microbiome. Am J Physiol Ren Physiol 316(6):F1211–F1217. https://doi.org/10.1152/ajprenal.00298.2018
Wong N, Wang X (2015) miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res 43(Database issue):D146–152. https://doi.org/10.1093/nar/gku1104
Agarwal V, Bell GW, Nam JW, Bartel DP (2015) Predicting effective microRNA target sites in mammalian mRNAs. Elife 4. https://doi.org/10.7554/eLife.05005
Li JH, Liu S, Zhou H, Qu LH, Yang JH (2014) starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res 42(Database issue):D92–97. https://doi.org/10.1093/nar/gkt1248
Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, Benner C, Chanda SK (2019) Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun 10(1):1523. https://doi.org/10.1038/s41467-019-09234-6
Acknowledgements
No applicable.
Funding
This research was funded by the National Key Research and Development Program of China (2023YFF1204100), Tianjin Natural Science Foundation (23JCZXJC00040), Non-profit Central Research Institute Fund of Chinese Academy of Medical Sciences (2021-12 M-C&T-B-080 to S.F.), PUMC Youth Fund and supported by the Fundamental Research Funds for the Central Universities of China (3332021055 to X.Z) and the National Natural Science Foundation of China (81670171 to E.J., 81601369 to X.P., 81870090 to S.F., 82101853 to W.S.), and the National Basic Research Program of China (2015CB964402 to E.J.).
Author information
Authors and Affiliations
Contributions
Conceptualization, X.P. and S.F.; methodology, L.L.,J.W., and Y.C.; software, Y.R.; validation, X.P., S.F. and W.S.; formal analysis, W.S.; investigation, J.L. and X.Z.; resources, S.Z.; data curation, E.J.; writing—original draft preparation, W.S.; writing—review and editing, Y.W. and W.S.; visualization, X.P.; supervision, X.P.; project administration, S.F.; funding acquisition, S.F. All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Informed consent statement
Informed consent was obtained from all subjects involved in the study.
Competing interests
The authors declare no competing interests.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Ethics Committee of Blood Diseases Hospital, Chinese Academy of Medical Sciences (Approval No. CAMSCRF2021013-EC-2, 2021-03-01) and the Ethics Committee of Tianjin First Central Hospital (Approval No. 2019N134KY, 2019-01-04).
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Appendix A
Appendix A
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Shao, W., Wang, Y., Liu, L. et al. Combining serum microRNAs and machine learning algorithms for diagnosing infectious fever after HSCT. Ann Hematol 103, 2089–2102 (2024). https://doi.org/10.1007/s00277-024-05755-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00277-024-05755-3