Abstract
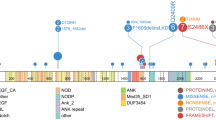
The molecular pathogenesis of extranodal NK/T-cell lymphoma (NKTCL) remains obscured despite the next-generation sequencing (NGS) studies explored on ever larger cohorts in the last decade. We addressed the highly variable mutation frequencies reported among previous studies with comprehensive amplicon coverage and enhanced sequencing depth to achieve higher genomic resolution for novel genetic discovery and comparative mutational profiling of the oncogenesis of NKTCL. Targeted exome sequencing was conducted to interrogate 415 cancer-related genes in a cohort of 36 patients with NKTCL, and a total of 548 single nucleotide variants (SNVs) and 600 Copy number variances (CNVs) were identified. Recurrent amplification of the MCL1 (67%) and PIM1 (56%) genes was detected in a dominant majority of patients in our cohort. Functional mapping of genetic aberrations revealed that an enrichment of mutations in the JAK-STAT signaling pathway, including the cytokine receptor LIFR (copy number loss) upstream of JAK3, STAT3 (activating SNVs), and downstream effectors of MYC, PIM1 and MCL1 (copy number gains). RNA in situ hybridization showed the significant consistence of MCL1 RNA level and copy number of MCL1 gene. We further correlated molecular and clinical parameters with overall survival (OS) of these patients. When correlations were analyzed by univariate followed by multivariate modelling, only copy number loss of LIFR gene and stage (III-IV) were independent prognostic factors of reduced OS. Our findings identified that novel loss of LIFR gene significantly correlated with the adverse clinical outcome of NKTCL patients and provided therapeutic opportunities for this disease through manipulating LIFR.






Similar content being viewed by others
References
Ren W et al (2017) Distinct subtype distribution and somatic mutation spectrum of lymphomas in East Asia. Curr Opin Hematol 24(4):367–376
Hu B, Oki Y (2018) Novel Immunotherapy options for Extranodal NK/T-Cell Lymphoma. Front Oncol 8:139
Martinez GS, Ross JA, Kirken RA (2016) Transforming mutations of Jak3 (A573V and M511I) show Differential Sensitivity to selective Jak3 inhibitors. Clin Cancer Drugs 3(2):131–137
de Mel S et al (2019) Molecular pathogenic pathways in extranodal NK/T cell lymphoma. J Hematol Oncol 12(1):33
Cai Q et al (2019) Epstein-Barr virus-positive natural Killer/T-Cell lymphoma. Front Oncol 9:386
Tse E, Kwong YL (2017) The diagnosis and management of NK/T-cell lymphomas. J Hematol Oncol 10(1):85
Zhang Y et al (2018) Frequent mutations in natural Killer/T cell lymphoma. Cell Physiol Biochem 49(1):1–16
Haverkos BM et al (2016) Extranodal NK/T Cell Lymphoma, nasal type (ENKTL-NT): an update on Epidemiology, Clinical Presentation, and natural history in north American and European cases. Curr Hematol Malig Rep 11(6):514–527
van Doesum JA et al (2021) Extranodal Natural Killer/T-cell lymphoma, nasal type: diagnosis and treatment. Hemasphere 5(2):e523
Jeong SH (2020) Extranodal NK/T cell lymphoma. Blood Res 55(S1):S63–S71
Au WY et al (2009) Clinical differences between nasal and extranasal natural killer/T-cell lymphoma: a study of 136 cases from the International Peripheral T-Cell Lymphoma Project. Blood 113(17):3931–3937
Schuler A et al (2017) Extranodal natural killer/T-cell lymphoma, nasal type: a rare but critical diagnosis. JAAD Case Rep 3(3):225–227
Saleem A, Natkunam Y (2020) Extranodal NK/T-Cell Lymphomas: the role of natural killer cells and EBV in Lymphomagenesis. Int J Mol Sci, 21(4)
Kim WY et al (2019) Epstein-Barr Virus-Associated T and NK-Cell Lymphoproliferative diseases. Front Pediatr 7:71
Vose J et al (2008) International peripheral T-cell and natural killer/T-cell lymphoma study: pathology findings and clinical outcomes. J Clin Oncol 26(25):4124–4130
Kim SJ et al (2009) Phase II trial of concurrent radiation and weekly cisplatin followed by VIPD chemotherapy in newly diagnosed, stage IE to IIE, nasal, extranodal NK/T-Cell Lymphoma: Consortium for Improving Survival of Lymphoma study. J Clin Oncol 27(35):6027–6032
Yamaguchi M et al (2009) Phase I/II study of concurrent chemoradiotherapy for localized nasal natural killer/T-cell lymphoma: Japan Clinical Oncology Group Study JCOG0211. J Clin Oncol 27(33):5594–5600
Tsai HJ et al (2015) Long-term results of a phase II trial with frontline concurrent chemoradiotherapy followed by consolidation chemotherapy for localized nasal natural killer/T-cell lymphoma. Eur J Haematol 94(2):130–137
Tse E, Au-Yeung R, Kwong YL (2019) Recent advances in the diagnosis and treatment of natural killer/T-cell lymphomas. Expert Rev Hematol 12(11):927–935
Dufva O et al (2018) Aggressive natural killer-cell leukemia mutational landscape and drug profiling highlight JAK-STAT signaling as therapeutic target. Nat Commun 9(1):1567
Vogler M, Walter HS, Dyer MJS (2017) Targeting anti-apoptotic BCL2 family proteins in haematological malignancies - from pathogenesis to treatment. Br J Haematol 178(3):364–379
Montes-Mojarro IA et al (2019) Mutational profile and EBV strains of extranodal NK/T-cell lymphoma, nasal type in Latin America. Mod Pathol
Li Z et al (2019) Recurrent GNAQ mutation encoding T96S in natural killer/T cell lymphoma. Nat Commun 10(1):4209
Gao LM et al (2019) Somatic mutations in KMT2D and TET2 associated with worse prognosis in Epstein-Barr virus-associated T or natural killer-cell lymphoproliferative disorders. Cancer Biol Ther 20(10):1319–1327
Sim SH et al (2017) Novel JAK3-Activating mutations in Extranodal NK/T-Cell Lymphoma, nasal type. Am J Pathol 187(5):980–986
Dobashi A et al (2016) Frequent BCOR aberrations in extranodal NK/T-Cell lymphoma, nasal type. Genes Chromosomes Cancer 55(5):460–471
Choi S et al (2016) Mutational analysis of Extranodal NK/T-Cell Lymphoma using targeted sequencing with a Comprehensive Cancer Panel. Genomics Inf 14(3):78–84
Lee S et al (2015) Genetic alterations of JAK/STAT cascade and histone modification in extranodal NK/T-cell lymphoma nasal type. Oncotarget 6(19):17764–17776
Kucuk C et al (2015) Activating mutations of STAT5B and STAT3 in lymphomas derived from gammadelta-T or NK cells. Nat Commun 6:6025
Jiang L et al (2015) Exome sequencing identifies somatic mutations of DDX3X in natural killer/T-cell lymphoma. Nat Genet 47(9):1061–1066
Xiong J et al (2020) Genomic and Transcriptomic Characterization of Natural Killer T Cell Lymphoma. Cancer Cell 37(3):403–419e6
Somasundaram N et al (2019) Pathogenesis and biomarkers of natural killer T cell lymphoma (NKTL). J Hematol Oncol 12(1):28
Karube K et al (2011) Identification of FOXO3 and PRDM1 as tumor-suppressor gene candidates in NK-cell neoplasms by genomic and functional analyses. Blood 118(12):3195–3204
Gao Y et al (2020) KMT2D and TP53 mutation status improve the prognostic value of the International Prognostic Index (IPI) stratification in ENKTL patients. Neoplasma
Xiong J, Zhao WL (2018) Advances in multiple omics of natural-killer/T cell lymphoma. J Hematol Oncol 11(1):134
Cho J et al (2020) Immune subtyping of extranodal NK/T-cell lymphoma: a new biomarker and an immune shift during disease progression. Mod Pathol 33(4):603–615
++ et al (2011) Activated oncogenic pathways and therapeutic targets in extranodal nasal-type NK/T cell lymphoma revealed by gene expression profiling. J Pathol 223(4):496–510
Huang Y et al (2010) Gene expression profiling identifies emerging oncogenic pathways operating in extranodal NK/T-cell lymphoma, nasal type. Blood 115(6):1226–1237
Coppo P et al (2009) STAT3 transcription factor is constitutively activated and is oncogenic in nasal-type NK/T-cell lymphoma. Leukemia 23(9):1667–1678
de Mel S et al (2018) The Genomics and Molecular Biology of Natural Killer/T-Cell Lymphoma: opportunities for translation. Int J Mol Sci, 19(7)
Wang L et al (2021) LncRNA BCYRN1-induced autophagy enhances asparaginase resistance in extranodal NK/T-cell lymphoma. Theranostics 11(2):925–940
Boeva V et al (2014) Multi-factor data normalization enables the detection of copy number aberrations in amplicon sequencing data. Bioinformatics 30(24):3443–3450
Wang Z et al (2013) Automated quantitative RNA in situ hybridization for resolution of equivocal and heterogeneous ERBB2 (HER2) status in invasive breast carcinoma. J Mol Diagn 15(2):210–219
Huang da W, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4(1):44–57
Koo GC et al (2012) Janus kinase 3-activating mutations identified in natural killer/T-cell lymphoma. Cancer Discov 2(7):591–597
Hanahan D, Weinberg RA (2011) Hallmarks of cancer: the next generation. Cell 144(5):646–674
Seeneevassen L et al (2020) Leukaemia inhibitory factor (LIF) inhibits Cancer Stem cells tumorigenic properties through Hippo kinases activation in gastric Cancer. Cancers (Basel), 12(8)
Johnson RW et al (2016) Induction of LIFR confers a dormancy phenotype in breast cancer cells disseminated to the bone marrow. Nat Cell Biol 18(10):1078–1089
Nicola NA, Babon JJ (2015) Leukemia inhibitory factor (LIF). Cytokine Growth Factor Rev 26(5):533–544
Luo Q et al (2015) LIFR functions as a metastasis suppressor in hepatocellular carcinoma by negatively regulating phosphoinositide 3-kinase/AKT pathway. Carcinogenesis 36(10):1201–1212
Chen D et al (2012) LIFR is a breast cancer metastasis suppressor upstream of the Hippo-YAP pathway and a prognostic marker. Nat Med 18(10):1511–1517
Dong G et al (2022) Genomic profiling identifies distinct genetic subtypes in extra-nodal natural killer/T-cell lymphoma. Leukemia 36(8):2064–2075
Acknowledgements
The authors thank Ms. Ling Huang for help with the tumor samples processing and DNA extraction. We thank Dr. Chi-Kuan Chen for the pathology review at MacKay Memorial Hospital. We also thank Drs. Shang-Yun Liu and Yi-Ting Yang (ACT genomics) for initial bioinformatic analysis of the NGS raw data.
Funding
This work was supported by a grant from the National Science and Technology Council, Taipei, Taiwan (NSTC 111-2314-B195-007) to Y.-C. C. and intramural funding from the Department of Medical Research, MacKay Memorial Hospital (MMH-110-75 and MMH-111-70) to Y.-C. C.
Author information
Authors and Affiliations
Contributions
Y.-C. C., H.-J. T., K.-H.L. and K.-C.C. conceived and designed the study; Y.-C. C., H.-J. T., N.-W.S., Y.-W.S., Y.-F.C., C.G.-S.C., J.L., M.-C.C., K.-H.L. and K.-C.C provided study materials or patient care; Y.-C. C., H.-J. T., T.-Y.H., N.-W.S., Y.-W.S., Y.-F.C., C.G.-S.C., J.L., M.-C.C., S.-J.C., H.-C.C., K.-H.L., K.-C.C. and S.-H.K. analyzed and interpreted the data; S.-J.C. and H.-C.C. reviewed the NGS raw data and initial analysis reports; Y.-C. C., H.-J. T.-Y.H., K.-H.L. and S.-H.K. wrote the manuscript; and all authors provided final approval of the manuscript. Conflict-of-interest disclosure Drs. Shu-Jen Chen and Hua-Chien Chen are employees of ACT Genomics Co., Ltd, Taipei, Taiwan. All the other authors declare no competing financial interests.
Corresponding authors
Ethics declarations
Disclosure
Drs. Shu-Jen Chen and Hua-Chien Chen are employees of ACT Genomics Co., Ltd, Taipei, Taiwan. All the other authors declare no competing financial interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chang, YC., Tsai, HJ., Huang, TY. et al. Analysis of mutation profiles in extranodal NK/T-cell lymphoma: clinical and prognostic correlations. Ann Hematol (2024). https://doi.org/10.1007/s00277-024-05698-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00277-024-05698-9