Abstract
Background
Molecular lymph node workup with one-step nucleic acid amplification (OSNA) is a validated diagnostic adjunct in breast cancer and also appealing for colon cancer (CC) staging. This study, for the first time, evaluates the prognostic value of OSNA in CC.
Patients and methods
The retrospective study includes patients with stage I-III CC from three centres. Lymph nodes were investigated with haematoxylin and eosin (H&E) and with OSNA, applying a 250 copies/μL threshold of CK19 mRNA. Diagnostic value of H&E and OSNA was assessed by survival analysis, sensitivity, specificity and time-dependent receiver operating characteristic curves.
Results
Eighty-seven patients were included [mean follow-up 53.4 months (± 24.9)]. Disease recurrence occurred in 16.1% after 19.8 months (± 12.3). Staging with H&E independently predicted worse cancer-specific survival in multivariate analysis (HR = 10.77, 95% CI 1.07–108.7, p = 0.019) but not OSNA (HR = 3.08, 95% CI 0.26–36.07, p = 0.197). With cancer-specific death or recurrence as gold standard, H&E sensitivity was 46.7% (95% CI 21.3–73.4%) and specificity 84.7% (95% CI 74.3–92.1%). OSNA sensitivity and specificity were 60.0% (95% CI 32.3–83.7%) and 75.0% (95% CI 63.4–84.5%), respectively.
Conclusions
In patients with CC, OSNA does not add relevant prognostic value to conventional H&E contrasting findings in other cancers. Further studies should assess lower thresholds for OSNA (< 250 copies/μL).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In the last decades, improvements in surgical techniques as well as in pathological assessment and adjuvant therapy could significantly enhance 5-year survival rates of patients with colon cancer. Positive lymph node (LN) status (stage III) is still one of the most important prognostic factors determining the allocation of adjuvant chemotherapy [1]. Most interestingly, there exists no real gold standard for reliable identification of negative and positive LN [2]. The standard histopathological LN workup consists of a haematoxylin and eosin (H&E) staining of one to two sections of each retrieved LN. Such conventional histopathological workup carries the substantial risk of “understaging” due to undetected micro- or macrometastases (i.e. tissue allocation bias). A minimum of 12 analysed LN is therefore recommended by most national cancer organizations [3, 4]. However, the agreement to yield a minimum of 12 LN was proposed over two decades ago and up to date clear evidence to justify this threshold is still missing [5, 6]. Furthermore, determination of a certain number of LN cannot compensate for a missing “gold standard” and the inherent tissue allocation bias. Understaging may therefore substantially contribute to the high recurrence rate (20–25%) of patients with negative LN status [7]. It was demonstrated that multilevel LN sectioning combined with immunohistochemistry can improve the detection rate of small nodal tumour infiltrates (i.e. isolated tumour cells and micrometastases), though it is a costly and protracted process [8]. Considering the possible tissue allocation bias as well as the cost and time intensive multilevel sectioning, molecular methods like one-step nucleic acid amplification (OSNA) are appealing since they do not require microscopic examination, and are standardized and fast. For OSNA, cytokeratin 19 (CK19) mRNA is amplified based on reverse transcription-loop-mediated isothermal amplification [9]. CK19 is one of the most widely used immunohistochemical marker to identify epithelial cells and is highly suggestive for the presence of colon cancer metastases when detected in a respective LN [10,11,12].
Up to date, only a few studies evaluated the clinical performance of OSNA in colon cancer. These studies compared OSNA with conventional pathological workup and immunohistochemistry techniques showing a high concordance level between the investigated methods [12,13,14,15,16]. However, the prognostic value of OSNA and thus its suitability as a “gold standard” to distinguish between LN positive and LN negative patients with colon cancer is unknown as respective data are entirely missing. The purpose of the here presented study was to evaluate the prognostic value of OSNA in patients with stage I-III colon cancer.
Patients and methods
For this retrospective study, patients with Union for International Cancer Control stage I, II and III colon cancer were identified from three centres—Cantonal Hospital Olten, Switzerland, Lausanne University Hospital, Switzerland, and Leiden University Medical Centre, Netherlands—that previously participated in three published cohort studies [13,14,15], except the patients recruited at the Lausanne University Hospital. Medical ethical approval was obtained from the local ethical committees, and the obtainment of patient consent is mentioned both in detail in the three previous publications [13,14,15]. The study was performed in accordance with the Declaration of Helsinki.
Included patients were over 18 years old with histological proven colon carcinoma, localized above the peritoneal fold, and submitted to in vivo (Olten [13], Lausanne) or submitted to ex vivo (Olten [14], Leiden [15]) sentinel LN mapping.
Recruitment occurred between January 2007 and December 2013. Follow-up data were retrieved from the patients’ medical files and treating general practitioners until October 2017.
Exclusion criteria were carcinoma in situ, stage IV colon cancer or preoperative neoadjuvant treatment.
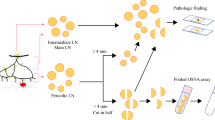
The study protocols of the previous cohort studies were in detail mentioned elsewhere [13,14,15]. In brief, sentinel LN mapping with isosulfan blue (Cantonal Hospital Olten and Lausanne University Hospital) or indocyanine green (Leiden University Medical Centre) was performed during standard oncological colon resection. Forty (22 [13] and 18 [14]) patients from Cantonal Hospital Olten, 21 from Lausanne University Hospital and 26 from Leiden University Medical Centre [15] were included. All harvested LN over 3 mm in greatest dimension [13] or a short axis ≥ 10 mm [15] was cut into 4 slices labelled from a to d. Blocks a and c were stored for later OSNA analysis. Blocks b and d were fixed in neutral buffered formaldehyde, processed in paraffin blocks and allocated to conventional standard H&E staining, multilevel H&E staining and immunohistochemistry (IHC) for CK19 as described elsewhere [13, 15].
In 18 of 40 patients from Cantonal Hospital Olten—the second study [14] – the harvested LN was processed cutting a middle section of 1 mm dedicated to standard H&E. The remaining parts of the LN were investigated by OSNA. Since the main focus of the present study was to evaluate the prognostic value of OSNA analysis, all patients from the three centres were included, despite the slightly different LN processing protocol.
For OSNA analysis, a CK19 mRNA copy threshold of 250 copies/μL was used. A CK19 mRNA copy number of < 250 copies/μL was considered negative for metastatic LN involvement, and a copy number ≥ 250 copies /μL was considered positive.
This was defined by previous studies using the logarithmic midpoint between the maximum value of the CK19 mRNA copy number in LN from pN0 patients and minus 2 standard deviations (SD) [12] or minus 3 SD [17] value from the average of CK19 mRNA copy number in histopathologically positive LN.
These studies also defined the micrometastases threshold between 250 and 4999 CK 19 mRNA copies/ μL. LN with 5000 or more mRNA copies/ μL was considered macrometastases. [12, 17].
As currently no “gold standard” for the definition of “real” negative and positive LN exists, we used the event of cancer-related death and cancer-related recurrence as proof of real node positivity and patients without disease recurrence or cancer-related death during follow-up were indexed to real negative LN.
The 30-day mortality and overall mortality were documented as well as the cause of death. Tumour recurrences were defined by radiological, endoscopic or histological evidence.
Statistical analysis
Statistical analysis was performed using the R environment (http://www.rproject.org). Two-sided p values < 0.05 were considered statistically significant. Continuous data were expressed as the mean ± standard deviation (SD). Confidence intervals (CIs) of binominal proportions were estimated as exact binomial 95% CI. After descriptive analysis of patient and tumour data for centre effects, the impact of detection of metastatic LN by H&E and OSNA on cancer specific survival (CSS), cancer-specific and recurrence-free survival (CSRFS) and disease-free survival (DFS) was assessed by Cox regression analysis with and without risk adjustment for patient and tumour data. Additionally, backward variable selection procedures from the full Cox regression model based on the Akaike's information criterion (AIC) were performed. The proportional hazard assumption was tested by scaled Schoenfeld residuals and by inspection of the hazard ratio (HR) plots. CI for hazard ratios (HR) were estimated with the Wald method. Significance tests were performed as likelihood ratio tests [18]. Then, sensitivity, specificity and diagnostic odds ratio for detection of metastatic LN by H&E and OSNA based on cancer-specific deaths or recurrence as the gold standard for true LN positivity were estimated. To further assess the diagnostic accuracy of H&E and OSNA and the potential influence of their cut-offs (≥ 250 copies /μL for OSNA and one positive LN for H&E), time-dependent receiver operating characteristic (ROC) curves were estimated from the censored survival data [19]. The area under the ROC curve (AUC) was computed using the nonparametric trapezoidal method [20]. Cut-off values were estimated by maximizing the Youden index.
Results
Patient and tumour characteristics
A total of 87 patients with stage I-III colon cancer were included in this retrospective study. Patient and tumour characteristics did not differ relevantly between the three centres (Table 1).
Follow-up and outcome
The mean follow-up time was 53.4 months (SD ± 24.9). A total of 14 patients (16.1%) developed disease recurrence, with a mean time to recurrence of 19.8 months (SD ± 12.3). Thirteen patients had distant metastases; one patient was diagnosed with a local recurrence. The overall mortality was 32.2% (28 patients) from which 10.3% (nine patients) died due to recurrent disease.
Survival analysis
In univariable analysis, detection of positive LN with H&E staining was associated with worse DFS (HR = 2.35, 95% CI 1.18–4.69, p = 0.019), CSS (HR = 10.44, 95% CI 2.18–49.94, p < 0.001) and CSRFS (HR = 6.67, 95% CI 2.30–19.31, p < 0.001). For OSNA, no impact on DFS (HR = 1.74, 95% CI 0.88–3.43, p = 0.123), worse CSS (HR = 7.69, 95% CI 1.65–35.76, p p = 0.004) and CSRFS (HR = 3.57, 95% CI 1.29–9.85, p = 0.015) were observed (Fig. 1 and Table 2).
Multivariate analysis confirmed detection of positive LN by H&E staining as a significant predictor for worse CSS (HR = 10.77, 95% CI 1.07–108.07, p = 0.019) and CSRFS (HR = 5.96, 95% CI 1.20–29.63, p = 0.017). For DFS (HR = 2.56, 95% CI 0.96–6.80, p = 0.064), a tendency for worse survival was observed. In contrast, no impact of OSNA positivity on DFS (HR = 0.93, 95% CI 0.40–2.21,p = 0.873) CSS (HR = 3.08, 95% CI 0.26–36.07, p = 0.197) and CSRFS (HR = 1.25, 95% CI 0.30–5.15, p = 0.727) persisted after multivariable adjustment (Table 2). The backward variable selection procedures uniformly confirmed the detection of positive LN by H&E but not by OSNA as significant predictors for CSS, DFS, and CSRFS.
Diagnostic accuracy of H&E and OSNA
Considering cancer-specific death or recurrence as the gold standard for true LN positivity, sensitivity was 46.7% (95% CI 21.3–73.4%) for H&E and 60.0% (95% CI 32.3–83.7%) for OSNA. Specificity was 84.7% (95% CI 74.3–92.1%) and 75.0% (95% CI 63.4–84.5%) for H&E and OSNA respectively. The diagnostic odds ratio of H&E was 485.2 (95% CI 146.1–1611.8) compared to 450 (95% CI 140.7–1439.2) for OSNA.
Limiting the analysis to patients with a minimum of 12 harvested LN, sensitivity of H&E and OSNA were identical (66.7%; 95% CI 22.3–95.7%), while specificity of H&E was 75% (95% CI 47.9–92.7) and of OSNA 68.8% (95% CI 41.3–86.1).
Time-dependent diagnostic accuracy of H&E and OSNA for survival prediction
Both H&E and OSNA had a low to moderate predictive value for DFS (AUC of 0.562 and 0.573), CSS (AUC of 0.740 and 0.600) and CSRFS (AUC of 0.661 and 0.630) (Fig. 2). According to the ROC curves in Fig. 2 the cut-off for H&E staining, set to a minimum of one positive LN, obtains the best diagnostic value. For OSNA, the ROC curves suggest the possibility of higher AUC with cut-off values lower than 250 mRNA copies/μL, which were not assessed in the present study (Table 3). The diagnostic value of H&E was higher compared to OSNA for CSS and CSRFS: the observed sensitivity (defined as the rate of death predicted and not survived) and specificity (defined as 1 minus the rate of patients with death predicted and survived) for H&E staining for 5-year DFS were 37.6% and 77.0%, for 5-year CSS 75.0% and 78.8%, and for 5-year CSRFS 57.7% and 78.0%. The observed sensitivity and specificity for OSNA for 5-year DFS were 38.8% and 72.8%, for 5-year CSS 51.2% and 70.4%, and for 5-year CSRFS 48.8% and 72.2%.
Time-dependent receiver operating characteristic (ROC) curves for 5-year survival rates. Curves for prediction by H&E staining are depicted in panels a, c, e and curves for prediction by OSNA in panel b, d, f. Panels a and b depict the disease-free survival, panels c and d depict the cancer-specific survival, and panels e and f depict the cancer-specific and recurrence-free survival. The area under the curve (AUC) is a measure for the diagnostic accuracy of the predictions with values of 1 representing a perfect prediction and values of 0.5 presenting a prediction by random. On the x-axis the false positives (death predicted, survived) and on the y-axis the true positives (death predicted, died) are shown. The true and false positives are estimated for each possible cut-off value (number of positive lymph nodes for H&E and Units for OSNA)
Discussion
This is the first study to evaluate the prognostic value of OSNA analysis in patients with colon cancer. According to the here presented results, OSNA offers no prognostic advantage compared to conventional LN staging with H&E. Interestingly, our previous studies identified OSNA as a promising diagnostic tool in colon cancer, showing high concordance rates with conventional histopathological workup with H&E, which were also confirmed by other authors in subsequent studies [13,14,15, 21]. However, all these previous trials investigated the performance of OSNA for the histopathological staging of colon cancer LN compared to either staging with H&E or multilevel sectioning and staining with immunohistochemistry and did not elucidate its prognostic value. Assessing the prognostic value of a method for LN staging is challenging as a reasonable gold standard for the definition of LN negativity, and positivity still does not exist. As a worldwide standard, one to two sections of each retrieved LN are analysed with H&E. Yet this workup entails a big risk of missing small nodal tumour infiltrates (i.e. isolated tumour cells and micrometastases), and this method is therefore not suitable as a gold standard against which a new diagnostic method can be tested for comparison. To compare OSNA with standard H&E workup, we therefore defined a gold standard for the definition of LN negativity and positivity. Assuming correctly staged node negative patients will not develop disease recurrence, we set the event of disease recurrence or cancer-related death as the gold standard to distinguish between true node negative and positive disease. With this distinct definition of true LN negativity and positivity, the diagnostic accuracy of H&E as well as of OSNA was markedly impaired compared to our previous reports as well as to a recently published systematic review [22]. This phenomenon occurs most likely due to missed small nodal tumour infiltrates at initial staging, suggesting that understaging is a persistent problem regardless of the technique used. The understaging is a relevant problem in the histopathological assessment of colon cancer, which was mentioned by our group previously, showing that multilevel sectioning with immunohistochemistry leads to a relevant upstaging of 15.4–26% of otherwise negatively classified patients [8, 23]. However, the prognostic impact of small nodal tumour infiltrates is still a matter of debate.
The previous studies focused on a dichotomic scale for LN positivity defining the threshold at 250 CK19 mRNA copies per µL, which corresponds to micrometastases, making sure that macro- and micrometastases and not isolated tumour cells are scored as metastatic LN involvement [10, 12,13,14,15, 17]. Since the present study is a retrospective analysis, the same threshold was considered.
The time-dependent ROC curves suggested the possibility of higher AUC with cut-off values lower than 250 mRNA copies/μL. This could be explained by the presence of isolated tumour cells. However, the previous studies did not characterize the CK 19 mRNA threshold corresponding to isolated tumour cells.
The amount of CK19 mRNA correlates with the size of the LN metastases making it feasible to calculate the total tumor load (TTL) of a specimen [21, 24]. These studies showed that the TTL correlates with the number of metastatic LN.
However, Aldecoa et al. observed that high-grade (G3) tumours or tumours with vascular invasion (V1) presented lower levels of TTL making it not a reliable prognostic tool for these specific pathologic features [24].
The multivariate cox analysis confirmed that a positive LN status with H&E (UICC stage III) was a predictor of worse CSS, as previously described in the literature [25, 26]. On the other hand, detection of positive LN with OSNA did not demonstrate to predict a worse CSS. This lack of significance may be due to the fact that fewer LN per patient were analysed with OSNA as only a part of a sentinel LN was used for OSNA analysis. In the case of H&E, all LNs were always examined by means of H&E, which is why there might be a higher chance of predicting recurrences. However, non-sentinel LN was only assessed with one to two H&E-stained sections, according to the worldwide standard. It would be necessary to fully analyse all LN of each patient by means of H&E and OSNA. This is technically not possible, as LN needed for H&E cannot be processed for OSNA and vice versa. However, by investigating the sentinel LN we set our focus on those LN having the highest probability of harbouring metastases. Nevertheless, we cannot ensure both LN fragments to harbour a metastasis, meaning that the metastasis could be found only in the fragment assigned to H&E analysis and not in the OSNA fragment and vice-versa.
The Kaplan–Meier survival analysis showed that a positive LN status with conventional H&E was predictive for worse CSS, DFS and CSRFS, in contrast to an OSNA-positive LN status.
We would like to acknowledge the limitations of the present study. Our sample size is relatively small which may result in reduced statistical power. Nevertheless, for the time being it is the largest published cohort of patients in the literature and the first study investigating the prognostic impact of OSNA in patients with colon cancer so far and includes patients from three different centres. As a cohort study, the presence of confounders is a possibility but the patients' characteristics of all considered study centres were homogenous, making a selection bias unlikely.
The present study is the first evaluating the prognostic value of OSNA in patients with colon cancer so far and includes patients from three different centres. OSNA failed to outperform conventional H&E workup regarding its prognostic value. Despite its relevance in other fields (breast cancer), the results of the here presented study do not support the adoption of OSNA alone in the routine staging of patients with colon cancer. Further studies should assess lower thresholds for OSNA (< 250 copies/μL), in order to detect isolated tumour cells increasing the prognostic value of this molecular detection method.
References
André T, de Gramont A, Vernerey D et al (2015) Adjuvant fluorouracil, leucovorin, and oxaliplatin in stage II to III colon cancer: updated 10-year survival and outcomes according to BRAF mutation and mismatch repair status of the MOSAIC study. J Clin Oncol 33:4176–4187
Ong MLH, Schofield JB (2016) Assessment of lymph node involvement in colorectal cancer. Word J Gastrointest Surg 8:179–192
American college of surgeons (2018) CoC Quality of Care Measures https://www.facs.org/quality-programs/cancer/ncdb/qualitymeasures. Accessed Mar 20, 2018
Labianca R, Nordlinger B, Beretta GD et al (2013) Early colon cancer: ESMO clinical practice guidelines. Ann Oncol 24:64–72
Li Destri G, Di Carlo I, Scilletta R et al (2014) Colorectal cancer and lymph nodes: the obsession with the number 12. World J Gastroenterol 20:1951–1960
Fielding LP, Arsenault PA, Chapuis PH et al (1991) Clinicopathological staging for colorectal cancer: an international documentation system (IDS) and an international comprehensive anatomical terminology (ICAT). J Gastroenterol Hepatol 6:325–344
Weixler B, Viehl CT, Warschokow R et al (2017) Comparative analysis of tumor cell dissemination to the sentinel lymph nodes and to the bone marrow in patient with non-metastasized colon cancer: a prospective multicenter study. JAMA Surg 152:912–920
Viehl CT, Guller U, Cecini R et al (2012) Sentinel lymph node procedure leads to upstaging of patients with resectable colon cancer: results of the Swiss prospective, multicenter study sentinel lymph node procedure in colon cancer. Ann Surg Oncol 19:1959–1965
Notomi T, Okayama H, Masubuchi H et al (2000) Loop-mediated isothermal amplification of DNA. Nucleic Acids Res 28:E63
Croner RS, Schellerer V, Demund H et al (2010) One step nucleic acid amplification (OSNA)—a new method for lymph node staging in colorectal carcinomas. J Transl Med 8:83
Yamamoto N, Daito M, Hiyama K et al (2013) An optimal mRNA marker for OSNA (one-step nucleic acid amplification) based lymph node metastasis detection in colorectal cancer patients. Jpn J Clin Oncol 43:264–270
Yamamoto H, Sekimoto M, Oya M et al (2011) OSNA-based novel molecular testing for lymph node metastases in colon cancer patients: results from a multicenter clinical performance study in Japan. Ann Surg Oncol 18:1891–1898
Güller U, Zettl A, Worni M et al (2012) Molecular investigation of lymph nodes in colon cancer patients using one-step nucleic acid amplification (OSNA)—a new road to better staging? Cancer 118:6039–6045
Croner RS, Geppert C-I, Bader FG et al (2014) Molecular staging of lymph node-negative colon carcinomas by one-step nucleic acid amplification (OSNA) results in upstaging a quarter of patients in a prospective, European, multicenter study. Br J Cancer 110:2544–2550
Vogelaar FJ, Reimers MS, van der Linden RLA et al (2014) The diagnostic value of one-step nucleic acid amplification (OSNA) for sentinel lymph nodes in colon cancer patients. Ann Surg Oncol 21:3924–3930
Taniyana K, Motoshita J, Sakane J et al (2006) Combination analysis of a whole lymph node by one-step nucleic acid amplification and histology for intraoperative detection of micrometastasis. Pathobiology 73:183–191
Tsujimoto M, Nabakabayashi K, Yoshidome K et al (2007) One-step nucleic acid amplification for intraoperative detection of lymph node metastasis in breast cancer patients. Clin Cancer Res 13:4807–4816
Grambsch PM, Therneau TM (1994) Proportional hazards tests and diagnostics based weighted residuals. Biometrika 81:515–526
Headertz PJ, Lumley T, Pepe MS (2000) Time-dependent ROC curves for censored survival data and diagnostic marker. Biometrics 56:337–344
DeLong ER, DeLong DM, Clarke-Pearson DL (1988) Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics 44:837–845
Yamamoto H, Tomita N, Inomata M et al (2016) OSNA-assisted molecular staging in colorectal cancer: a prospective multicenter trial in Japan. Ann Surg Oncol 23:391–396
Wild JB, Iqbal N, Francombe J et al (2017) Is it time for one-step nucleic acid amplification (OSNA) in colorectal cancer? A systematic review and meta-analysis. Tech Coloprocto 21:693–699
Weixler B, Rickenbacher A, Raptis DA et al (2017) Sentinel lymph node mapping with isosulfan blue or indocyanine green in colon cancer shows comparable results and identifies patients with decreased survival: a prospective single center trial. World J Surg 41:2378–2386
Aldecoa I, Atares B, Tarragona J et al (2016) Molecularly determined total tumour load in lymph nodes of stage I-II colon cancer patients correlates with high-risk factors. A Multicentre Prospective Study. Virchows Arch 469:385–394
Merkel S, Mansmann U, Papadopoulos T et al (2001) The prognostic inhomogeneity of colorectal carcinomas stage III: a proposal for subdivision of stage III. Cancer 92:2754–2759
Chang GJ, Rodrigues-Bigas MA, Skibber JM et al (2007) Lymph node evaluation and survival after curative resection of colon cancer: systematic review. JNCI 99:433–441
Funding
Open Access funding provided by Universität Basel (Universitätsbibliothek Basel).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors disclaim no conflict of interest.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Weixler, B., Teixeira da Cunha, S., Warschkow, R. et al. Molecular Lymph Node Staging with One-Step Nucleic Acid Amplification and its Prognostic Value for Patients with Colon Cancer: The First Follow-up Study. World J Surg 45, 1526–1536 (2021). https://doi.org/10.1007/s00268-020-05949-6
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00268-020-05949-6