Abstract
Social structure plays a crucial role in determining a species’ dispersal patterns and genetic structure. Cetaceans show a diversity of social and mating systems, but their effects on dispersal and genetic structure are not well known, in part because of technical difficulties in obtaining robust observational data. Here, we combine genetic profiling and GIS analysis to identify patterns of kin distribution over time and space, to infer mating structure and dispersal patterns in short-beaked common dolphins (Delphinus delphis). This species is highly social, and exhibits weak spatial genetic structure in the Northeast Atlantic and Mediterranean Sea, thought to result from fluid social structure and low levels of site fidelity. We found that although sampled groups were not composed of closely related individuals, close kin were frequently found in the same geographic location over several years. Our results suggest that common dolphin exhibits some level of site fidelity, which could be explained by foraging for temporally varying prey resource in areas familiar to individuals. Dispersal from natal area likely involves long-distance movements of females, as males are found more frequently than females in the same locations as their close kin. Long-distance dispersal may explain the near panmixia observed in this species. By analysing individuals sampled in the same geographic location over multiple years, we avoid caveats associated with divergence-based methods of inferring sex-biased dispersal. We thus provide a unique perspective on this species’ social structure and dispersal behaviour, and how it relates to the observed low levels of population genetic structure in European waters.
Significance statement
Movement patterns and social interactions are aspects of wild animal’s behaviour important for understanding their ecology. However, tracking these behaviours directly can be very challenging in wide-ranging species such as whales and dolphins. In this study, we used genetic information to detect how patterns of kin associations change in space and time, to infer aspects of movement and social structure. We identified previously unknown site fidelity, and suggested that dispersal usually involves females, travelling long distances from the natal area. Our data analysis strategy overcomes known limitations of previously used genetic inference methods, and provides a new approach to identify differences in dispersal between the sexes, which contribute to better understanding of the species’ behaviour and ecology. In this case, we suggest that females are more likely to disperse than males, a pattern unusual amongst mammals.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In social animals, patterns of social interactions and kin association are determined by a combination of intrinsic and extrinsic factors, such as intra-specific competition, dispersal behaviour, predator avoidance and food availability. Social structure and dispersal patterns can also be linked with specific mating strategies, often resulting in sex-biased dispersal (Clutton-Brock and Lukas 2012). Therefore, insight into the interactions between kin association patterns, social structure and mating systems of wild animals can provide important information on their ecology, demographic structure and dispersal behaviour.
In highly mobile species that can travel over large distances, dispersal patterns are difficult to assess directly. Although individual movements can be tracked through tagging, radiotelemetry or satellite telemetry, they do not provide information on population-level dispersal patterns. Alternatively, this information can be inferred from patterns of genetic variability, assessed at the level of social groups and entire populations. For example, genetic studies of grey wolf (Canis lupus) populations revealed that dispersal of individuals is biased towards habitats similar to their natal habitats (Pilot et al. 2006; Musiani et al. 2007), which was previously unknown despite extensive ecological research on this species. Further, genetic studies on primates revealed that dispersal is mostly male-biased, with some exceptions such as chimpanzees (Pan troglodytes), bonobos (Pan paniscus) and hamadryas baboons (Papio hamadryas), where dispersal is female-biased (reviewed in Vigilant and Guschanski 2009).
Knowledge on dispersal patterns, mating systems and composition of social groups in cetaceans is limited due to difficulties with obtaining robust behavioural data in the marine environment. In several well studied species, such as bottlenose dolphins (Tursiops spp.) and killer whales (Orcinus orca), this knowledge has been obtained mostly from population genetic studies (reviewed in Möller 2012). All species of toothed cetaceans are known to form social groups, often grouping for the purposes of migration, protection, feeding and reproduction (Connor 2000). The size and social structure of these groupings can vary greatly between species (reviewed in Möller 2012) and in some cases even within species, as seen in killer whales and bottlenose dolphins (Ford et al. 2000; Connor and Krützen 2015). Such differences are thought to be determined by a combination of factors, including the availability of predictable prey resources (Möller 2012) and the risk of predation in open-water environments (Gowans et al. 2007). These are thought to affect patterns of kin association, but information is still lacking for many cetacean species (Möller 2012).
In some odontocetes, social groups tend to consist of individuals sharing close kinship relationships. Matrilineal pods, where a breeding female retains close association with its offspring, are seen in resident killer whales (O. orca; Ford et al. 2000), short-finned pilot whales (Globicephala macrorhynchus; Heimlich-Boran 1993) and female sperm whales (Physeter macrocephalus; Lyrholm and Gyllensten 1998). However, in other cetacean species such as Commerson’s dolphins (Cephalorhynchus commersonii; Coscarella et al. 2011) or Tucuxi (Sotalia guianensis; Santos and Rosso 2008), such close kin associations are not always present or occur only transiently (Connor 2000).
The social behaviour of bottlenose dolphins (Tursiops spp.) is a well-documented example of this type of social structure. This genus is characterised by a fission-fusion structure, where factors such as age, sex and reproductive status likely influence the social strategies of each individual dolphin (Connor 2000). Relatedness has also been identified as a factor, with female Tursiops aduncus in Eastern Australia showing preferred association with related females (Möller et al. 2006; Wiszniewski et al. 2010), whilst Tursiops truncatus in the Bahamas display greater frequency of male alliances between related individuals than would be expected by chance (Parsons et al. 2003). In spite of these well described cases, such effects of kinship on social association patterns have not been universally observed across populations of Tursiops spp. (Connor 2000; Möller 2012).
Common dolphins (Delphinus spp.) are highly social and are often seen in groups ranging from less than 30 to thousands of individuals (Perrin 2009). Information on dispersal patterns and site fidelity is extremely scarce, although genetic structure suggests potential long-range movements (Moura et al. 2013), supported by a single photo-ID observation (Genov et al. 2012). Scientific literature regarding its social structure and mating system is also scarce, with an earlier study suggesting it to be matrilineal (Amos 1999). More recently, a fission-fusion structure similar to bottlenose dolphins has been described (Bruno et al. 2004; Zanardo et al. 2016), which is consistent with the expectations for small pelagic delphinids (Möller 2012). Relative testis size (Murphy et al. 2005) indicates strong sperm competition, which is consistent with a more fluid social structure and low kinship association.
Short-beaked common dolphins in the Northeast Atlantic show low levels of genetic structure (Natoli et al. 2006; Mirimin et al. 2009; Moura et al. 2013), which also suggests limited kin clustering and high levels of gene flow. On a global scale, significant differentiation was found between populations inhabiting different ocean basins (Natoli et al. 2006), which was attributed to isolation by distance (Amaral et al. 2012). In certain regions of the eastern and southern Australian coasts, short-beaked common dolphins display fine-scale genetic structure (Bilgmann et al. 2008, 2014; Möller et al. 2011), which may imply differences in kin association patterns between regional populations.
A genetic study on kinship levels between individuals from a single mass stranding of common dolphin (Delphinus delphis) showed no evidence for kinship structure within a group (Viricel et al. 2008). However, mass-stranding events may not be representative of the natural social grouping of the species. In fact, a study on pilot whales (a species for which mass strandings are more frequent than for common dolphin) has revealed that the social structure inferred from such events differs strongly from that inferred from free-ranging groups in this species (Oremus et al. 2013).
Assessing kinship relations of common dolphins in their natural environment is thus required to gain new insight into the social organisation of the species. The frequency of first-, second- and third-degree relatives within social groups found in the wild may be used to further clarify common dolphin social structure and dispersal patterns. A recent study found that kinship could be an important determinant in group composition in Australian short-beaked common dolphin, particularly between males (Zanardo et al. 2016). Because kin association in dolphins can be transient, data from different time periods and locations are required to avoid context-dependent bias.
In this study, we provide an assessment of social structure in short-beaked common dolphin (D. delphis) based on free-ranging animals along the Portuguese coast, which are part of the larger Atlantic population where no studies on social structure have been carried out. Biopsy samples were obtained from multiple groups found in the same locations during multiple years, and kinship patterns were identified using individual multilocus genotypes. Kinship patterns were then analysed in the context of group composition, geographic location and year of sampling. We specifically assessed whether (1) groups of interacting common dolphin individuals are composed of related individuals, (2) individuals are found in the same locations as their close kin over multiple years and (3) kinship level between individuals increases with geographic proximity of locations where they are observed.
If the lack of genetic differentiation in European common dolphins is due to high promiscuity, with an associated fluid social structure and low philopatry, then no geographical patterns of kinship should be detected. Therefore, analysing the spatial and temporal distribution of kinship associations will allow us to not only improve our understanding of common dolphin social structure and dispersal patterns, but also shed light on how social structure shapes genetic structure in wild mammals. It will also evaluate the potential of genetic-based kinship analyses in inferring demographic and social patterns from elusive wild animals, for which field data might be difficult to obtain.
Methods
Sampling of wild dolphins
A total of 204 biopsy samples were collected from free-ranging, short-beaked common dolphins (D. delphis) in five separate locations along the Portuguese coast, separated by approximately 20 to 100 nautical miles. Samples were collected throughout three separate field seasons (2007–2009), although not all locations were sampled in all of the seasons (Table 1). Sampling was carried out with permits from the Portuguese Institute for the Conservation of Nature (ICNB) as described in (Moura et al. 2013). Importantly for this study, only animals that were clearly of adult size were sampled, and no animals accompanied by calves were targeted, so no bias in relatedness estimates is introduced from sampling mother-calf pairs. We cannot exclude that sexually immature individuals of adult size could have been collected alongside sexually mature adults. Sub-adults may display different kin association patterns as compared to adults (Mason et al. 2016), but in this study, it was not possible to assess age-specific kin structure.
We considered all individuals sampled in the same sampling event as belonging to the same group, irrespectively of the behaviour exhibited. A new sampling event was considered whenever more than 1 h had elapsed from the last biopsy sample taken from the previous group. This was the only criterion used to define groups, given that lack of knowledge of the social behaviour typical of this species locally, precluded accurate identification of social groups if sampling was interrupted for longer. Sampled groups ranged from approximately 20 to several hundred individuals, but exact numbers for each sampling event were not recorded. The sampling permit introduced a limit of ten samples per group to minimise animal welfare impact from the biopsy sampling effort. Within this limitation, each group was sampled as inclusively as possible to allow assessment of kinship within groups and to avoid bias from sampling potential nursing groups. It was not possible to record data blindly because our study involved focal animals in the field.
Kinship analyses
All individuals were genotyped for 15 microsatellite loci as described previously (Moura et al. 2013), and sex was determined using the ZFX/ZFY protocol (Bérubé and Palsbøll 1996). Estimating kinship from microsatellite genotypes alone may lead to misclassification of kinship relationships, if based on a small number of loci or if the loci analysed have low genetic variability (Blouin et al. 1996). However, moderate numbers (12–20) of highly variable loci provide sufficient resolution to be used as legal evidence (e.g. in assignment of human paternity or in individual identification) and were shown to accurately identify known kinship relationships in wild animals (e.g. Pilot et al. 2010b). To assess whether levels of genetic variation in our study population were high enough to achieve sufficient accuracy of kinship estimates, observed and expected heterozygosity were estimated for each locus individually and for overall loci, as well as for average individual heterozygosity, all calculated using GENALEX (Peakall and Smouse 2006). Furthermore, probability of identity as well as probability of no exclusion of parents and siblings were calculated in CERVUS (Kalinowski et al. 2007).
We identified two samples, collected in the same location but in different years, with an identical genotype, implying that the same individual was sampled twice. We kept the genotype from both samples when we assessed kinship relationships within groups, but removed the genotype of one sample for the comparisons between groups.
Individual’s parentage and kinship was estimated using the maximum-likelihood approach, which accounts for genotyping errors, implemented in the software packages CERVUS and KINGROUP (Konovalov et al. 2004). Because no prior information on social structure is available for the study population, only adults were sampled and their age was unknown; all individuals were considered as both candidate parents and offspring. Therefore, we applied a rigorous data analysis strategy to minimise the chances of obtaining false positives, as described in earlier studies (Pilot et al. 2010a; Moura et al. 2014).
Three parentage tests were performed in CERVUS: one to identify potential mother-offspring pairs, considering only the females as potential parents; another to identify potential father-offspring pairs, considering only the males as potential parents; and a last one to identify parent pairs, where males and females were considered as potential parents simultaneously. Confidence levels were estimated by simulating populations with known parent-offspring pairs, based on allele frequencies of the real population genotypes. Simulations assumed 10,000 offspring, 83 mothers and 122 fathers, corresponding to the number of males and females in the dataset (they were all adults and therefore potential parents). Simulations also assumed that 80% of parents of both sexes remain unsampled. We considered two confidence levels—strict (95%) and relaxed (80%).
In some cases, the parent-offspring pair was identified twice with individual parent-offspring assignment reversed. Because we did not know the age of individuals, it was impossible to know which of these assignments was correct, but for our purposes, it was not necessary to know which individual was a parent and which was an offspring. When such reverse parent-offspring assignments occurred, only one of the results was kept and the match noted. If no parentage was assigned to an individual at either strict or relaxed confidence level, then it was concluded that no parents or offspring of a given individual were sampled.
KINGROUP was used as a complementary method of identifying groups of close kin. We used this program to identify the most likely partition of all individuals into groups of kin related at a given level (e.g. full-siblings, half-siblings). The full sibship reconstruction (FSR) method was implemented, which partitions the analysed sample set into groups of kin sharing a common relationship. This allows for single-individual groups, which represent those individuals that do not have any relatives at the particular level (e.g. full-siblings), within the dataset. The most likely partition into kin groups is determined by calculating the likelihood that individuals in a group share a hypothesised kinship relationship (the primary hypothesis). Simulations are then used to test if that relationship level is significantly more likely than an alternative relationship (the null hypothesis). Because multiple alternative relationships are often possible, complex null hypotheses were tested. The primary hypothesis that the group consists of full-siblings was tested against the null hypothesis of the relationship at the level of half-siblings to unrelated. The primary hypothesis that the group consists of half-siblings was tested against the null hypothesis of the relationship at the level of cousins to unrelated.
Using this method, we reconstructed the partition of the analysed dataset into three kinship classes—full-siblings, half-siblings and cousins. KINGROUP has a limited ability of distinguishing between parent-offspring and full-siblings, so CERVUS parent-offspring pairs were accepted if KINGROUP found them to be related at either a parent-offspring or a full-sibling level.
Because KINGROUP identifies groups of individuals at a particular level of relatedness, full-sibling groups may also include parent-offspring pairs (with the same expected proportion of 50% shared alleles), and half-sibling groups may include, e.g. grandparent-offspring pairs (with the expected proportion of 25% shared alleles). Parent-offspring pairs were distinguished from full-sibling pairs by including an additional condition that they have to share at least one allele at each locus (full-siblings share 50% alleles at average, but may not share any alleles at some individual loci).
When comparing the two parents-offspring trio matches to establish the likelihood of an assigned parent being correct, the number of mismatching loci and pair confidence were considered. The putative parent with the least mismatching loci and an assigned confidence level was predicted to be the most likely parent.
Patterns of kin group occurrence and distribution
Kin relationships were then integrated with information regarding sampling group, time of sampling and location of sampling. Specifically, we were interested in determining which of those factors maximised the grouping of related individuals. It should be noted that because of how groups were defined (see earlier), individual groups might have been sampled multiple times in the same day, if found at discrete times of the day.
We determined the number of kin groups including individuals sampled in the same day, month and year for each of the three different kinship relationships independently, and compared them to groups containing individuals sampled across different years. The number of kin groups containing individuals sampled exclusively in the same location, was also compared to the number of kin groups containing individuals sampled in different locations. When a group was found to be sampled only in two different locations, we discriminated whether these locations were geographically next to each other or not. Correlation between the number of kin groups and the location was assessed through contingency table analyses, using a Pearson chi-squared test and a Fisher exact test (in tables small enough for computations) using the software PAST (Hammer et al. 2001). These analyses were repeated separately for males and females in the parent-offspring category to test for potential sex-biased dispersal from natal group. Further tests for sex-biased dispersal were performed in the FSTAT package (Goudet 2001; Goudet et al. 2002) by comparing the sex-specific mean Assignment Index (mAIc), variance of Assignment Index (vAIc), pairwise F ST between locations, mean F IS and mean pairwise relatedness. If sex-biased dispersal is occurring, then the dispersing sex is expected to have lower mAIc, F ST, F IS and mean pairwise relatedness, but higher vAIc.
For cousin groups inferred in KINGROUP, we calculated the proportion of each group that was found in a given location, and represented the proportion geographically. This was done by defining the four points representing the maximum extent of sampling at each location, and performing ordinary krigging interpolation for each cousin group proportion at a given location, as implemented in the ArcMap (ESRI) geographic information system. This was done only for cousin groups, which reflect wider family relationships and therefore are more appropriate to assess the population level extent of natal dispersal. Furthermore, kin groups of closer relationships were usually composed of too few individuals to allow robust interpolation.
Results
Parentage and kinship analysis
In this study, parentage and kinship in the sampled population were inferred solely based on individual microsatellite genotypes, which require high levels of genetic variation (see “Methods” section for more details). In the population studied here, the probability of identity based on the 15 microsatellites was very low (PI = 7.23 × 10−18), as were all probabilities of non-exclusion of parents/siblings (all below 1.3 × 10−4). The heterozygosity levels across all loci were high (H E = 0.72, H O = 0.72, H Ind = 0.72), consistent with the heterozygosity levels described in other studies for common dolphin in the eastern North Atlantic (H E = 0.76–0.82, H O = 0.69–0.74, Natoli et al. 2006; H E = 0.705, H O = 0.695, Mirimin et al. 2009), though note that not all the same loci were used across studies. Two of the 15 microsatellite loci scored had H E lower than 0.2 (KWM1b and TexVet9). After excluding those two loci, mean H E and H O were both 0.81. Both values (0.72 and 0.81) are within the ranges identified as giving low rates of kinship misclassification (~5%) in earlier simulation studies (Blouin et al. 1996). Furthermore, although early methods of kin identification were based on estimating pairwise relatedness without assessing confidence levels of inferred relationships (Blouin et al. 1996; Van Horn et al. 2008), here we used methods of parentage and kinship analysis that estimate the likelihood of each inferred relationship. This provides an efficient way to minimise the occurrence of misclassified relationships.
The maximum-likelihood approach implemented in CERVUS 3.0 (Kalinowski et al. 2007) assigned a single parent to 68 individuals overall, with 39 being fathers and 29 being mothers. For 12 individuals, CERVUS assigned both parents. In addition, 30 other parent-offspring matches consisted of 15 pairs of matches where it was impossible to determine which individual was the parent and which was the offspring (as both relationships were assigned high confidence). Of all the assigned parents, three females were assigned as mothers to more than one offspring, and four males were assigned as fathers of two or more offspring.
Of all single parent assignments, seven fathers and six mothers were assigned with strict confidence (95%), whilst 25 fathers and 21 mothers were assigned with relaxed confidence level (80%). Of all parent-pair assignments, two pairs were assigned with strict confidence, and eight pairs were assigned with relaxed confidence. Although pairs assigned with relaxed confidence are more likely to be false positives, they are likely to represent close kin relationship other than parent-offspring pairs, such as full-siblings.
KINGROUP inferred a total of 22 full-sibling groups, 65 half-sibling groups and 25 cousin groups. Comparison with CERVUS results allowed six out of the 22 full-sibling pairs to be identified as parent-offspring matches (Table S1). Full-sibling groups consisted of two individuals, with half-sibling and cousin groups ranging from 2 to 4 and 2 to 19 individuals respectively, with 47% of half-sibling groups consisting of only two individuals (Fig. 1).
Temporal distribution of kin groups
Overall, our study sampled 46 different groups of interacting individuals, with an average of 3.9 sampled individuals per group (see Fig. S1 for the frequency distribution of group sample number). Individuals with close kinship relationships (parent-offspring, full-siblings and half-siblings) were never sampled in the same group, and 14 pairs of individuals related at the cousin level were sampled in the same group. No offspring were sampled on the same day or month as either of their parents, but they were frequently sampled in the same year (Table 2). On the other hand, one pair of full-siblings and several groups of half-siblings and cousins, included individuals that were sampled on the same day (Table S2). Regardless, most relatives were sampled in different years. For example, only 40% of the full-sibling groups were sampled in the same year (Table 2). Although the majority of half-sibling and cousin groups were composed of individuals sampled across different days/months, they often included individuals that were sampled in the same day/month. For example, cousin group 13 contained two individuals sampled on 28 August 2008, two sampled on 06 August 2009 and another two on 16 August 2009 (Table S2). The half-siblings and cousins sampled together at the same time, were all from kin groups larger than two individuals. This means that other members of the kin group were sampled in different social groups, and in many cases, they were sampled in different months or years.
Geographic distribution of kin groups
Overall, 16 parent-offspring pairs (as identified in CERVUS) were sampled in the same location (Table 3). The remaining 52 parent-offspring pairs were found in multiple locations, in different combinations between the six locations. For parent-offspring pairs, no mothers and only four fathers were sampled in the same location as the offspring, whilst none of the mother-father pairs were sampled in the same location (Table 3).
Generally, kin groups that were composed of more individuals tended to have a larger geographical area where members of the kin group were sampled (Table S2). Only three of the 31 half-sibling groups consisting of two individuals were sampled in the same area. Half-sibling and cousin groups often consisted of more than two individuals, and therefore, their distribution could be spread out over multiple locations.
Most kin groups identified in this study (independent of their size) were found in two or less locations (across all years); these could be either two neighbouring locations or two separate locations (i.e. locations that were not situated next to each other). Correlation between the number of kin groups and sampling location was significant (χ 2 = 18.233, p = 0.019), with most kin groups at a level higher than half-siblings (53%) found in the same or neighbouring locations relative to separate locations (although the difference was higher for father-offspring groups; Table 4).
When these analyses were repeated for parent-offspring pairs considering male and female offspring separately, a significant correlation was found between the number of kin groups and the sampling location for male offspring (χ 2 = 10.446, p = 0.033; Fisher’s p = 0.0297) but not for female offspring (χ 2 = 5.045, p = 0.283; Fisher’s p = 0.24). Fewer mothers were found in the same region as their offspring compared to fathers and their offspring (six vs ten pairs, respectively). Consistently, less female offspring were found in the same location as one of their parents than male offspring (six vs ten, respectively). For male offspring, most individuals were found in the same or neighbouring locations as their fathers, but in separate locations to their mothers (Table 4).
More full-sibling groups were found in separate locations than in the same or neighbouring locations (Table 4). Of those found in the same locations, more brothers and brother-sister pairs were sampled in the same location, compared with sister pairs. More brothers were sampled in the same location, whilst more sisters were sampled in neighbouring locations. However, these differences are marginal, as very few full-sibling groups were inferred. Amongst half-siblings and cousins, there were groups that included both individuals sampled in the same locations and individuals sampled in separate locations. In the case of half-siblings, four groups contained individuals that were all sampled in the same area (Table 4).
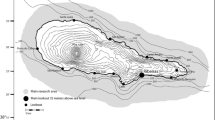
Only one group of cousins had members which were found in all sampled areas. Cousin groups were more commonly split between four areas (40% of all cousin groups); however, the geographical distribution of these groups shows that even in such cases, most individuals will be concentrated in only one or two areas (Fig. 2; e.g. groups 1, 2 and 10). These areas were often contiguous, but some cousin groups were separated by large geographic distances, which might include one or more locations where no members of that group were found (Fig. 2; e.g. groups 7, 8, 15 and 17). All classic sex-biased dispersal tests carried out were non-significant (Table S3), which would suggest no sex-biased dispersal, though note the discussion below regarding the applicability of such tests to this dataset.
Spatial distribution of biopsy samples and cousin groups of short-beaked common dolphins (Delphinus delphis) along the Portuguese coast. Numbers in brackets represent the sample size for each location. Yellow colour represents a high proportion of all individuals from that particular group, whilst blue colour represents no individuals from the group. Numbers 1–25 refer to different inferred cousin groups. Maps are not to scale; however, distance between the centres of Sagres and Portimão sample points is approximately 35 km
Discussion
Common dolphin social structure
The results of this study show that common dolphin social groups do not consist of closely related individuals, which is consistent with a previous study on mass strandings (Viricel et al. 2008). This contrasts with the kin-based social structure seen in other delphinids such as killer whales (Ford et al. 2000; Pilot et al. 2010b) or pilot whales (Heimlich-Boran 1993). The pattern observed in the common dolphin fits the expected pattern for small delphinids, where association of unrelated individuals is expected to occur due to reliance on unpredictable prey resources, as in such conditions there would be no benefit for either sex to exhibit any degree of philopatry (Möller 2012).
However, social groups that do not consist of close kin are relatively rare in mammals, and even in cases where social grouping is not based on kin association, such as gorillas, sub-groups of close kin within a group can be detected (Bradley et al. 2007). It cannot be excluded that the common dolphin groups also contain sub-groups of close kin, which remain undetected because our sample size was small relative to the size of the sampled groups. The geographic distribution of kin groups observed in our study suggests a certain degree of natal philopatry in common dolphins, with most groups containing kin found in the same or neighbouring location. This is consistent with a fission-fusion structure (Bruno et al. 2004), particularly such as reported for bottlenose dolphin (Connor and Krützen 2015). It should be noted that, given the short distance between sampling locations relative to the species’ dispersal abilities and known genetic panmixia across Europe for this species (Moura et al. 2013), groups found in neighbouring locations still likely reflect fidelity to a large home range, rather than dispersal from natal area.
The spatial distribution of kin inferred in our study differed between the sexes. Male kin were mostly found in the same or neighbouring locations (implying limited male dispersal), but female kin were mostly found in non-neighbouring, geographically distant locations, indicating that females disperse over longer distances than males on average. This result is consistent with those found in Zanardo et al. (2016), where kinship between individuals within a pod in common dolphins was higher for males than females.
Although our kinship analysis suggests a degree of female-biased dispersal in Northeast Atlantic common dolphins, other methods used gave non-significant results, which is consistent with previous results for this species on a larger scale (Natoli et al. 2008). However, methods based on sex-specific F statistics and assignment values, have low power in systems where the sex dispersal asymmetry is low and overall dispersal rates are high (Goudet et al. 2002). Common dolphins in Europe are panmictic (Moura et al. 2013), which results in very low pairwise F ST values between locations. Therefore, genetic differences between immigrants and phylopatric individuals will be low, which greatly reduces the power of classical sex-biased dispersal tests (Goudet et al. 2002). Therefore, we suggest that comparison of kin groups over space and time remains, for the moment, the most accurate method to assess sex-biased dispersal in European common dolphins, where sex-specific asymmetries in dispersal are likely to be subtle.
Potential drivers of common dolphin kinship and dispersal patterns
Inbreeding avoidance has been proposed as a potential mechanism promoting sex-biased dispersal (Perrin and Mazalov 2000; Lawson Handley and Perrin 2007; Clutton-Brock and Lukas 2012). The lack of association between parents and offspring found in this study may result from inbreeding avoidance in groups with a fluid social organisation. Female-biased dispersal as an inbreeding avoidance mechanism is more likely to develop in polygynous mating systems (Clutton-Brock 1989; Perrin and Mazalov 1999), but there is no evidence for common dolphins having such a mating system, and this is not supported by our results either.
Although fission-fusion social structures involve the creation of male alliances to maintain access to females in some bottlenose dolphin populations (Connor et al. 1992; Möller et al. 2001; Parsons et al. 2003), this has not been reported for common dolphins. Furthermore, relatively large testis size (Murphy et al. 2005; MacLeod 2010) suggests that females copulate with several males. Field observations are consistent with this, as during sampling efforts, we commonly observed a mating behaviour where a female allows several competing males to follow her until one male is chosen for copulation, although such behaviours have not been described in other small delphinid species.
It is thus likely that females mate with multiple males, in which case the mating system in common dolphins might be better described as polygynandrous (Watts 1998; Díaz-Muñoz et al. 2014). In such mating systems, sex-biased inbreeding avoidance strategies are more likely to result in male-biased dispersal (Dobson 1982; Lawson Handley and Perrin 2007). Given that the evidence presented here is more consistent with a certain degree of female-biased dispersal, we instead suggest that sex-biased dispersal observed in this study may reflect local ecological factors specific to this population.
Male-biased dispersal appears to be the norm in mammals (Greenwood 1980; Lawson Handley and Perrin 2007) and is largely thought to represent their ancestral state (Clutton-Brock and Lukas 2012). Nevertheless, cases of female-biased dispersal have also been described in mammalian species, such as the greater sac-winged bat (Nagy et al. 2007) and porcupines (Sweitzer and Berger 1998). For some species, such as the red deer, both male- (Catchpole et al. 2004) and female-biased (Pérez-González and Carranza 2009) dispersal have been suggested. In cetaceans, male-biased dispersal has been inferred only in species that inhabit environments where prey resources are predictable (Möller 2012), whilst species living in unpredictable environments have symmetrical sex dispersal. This includes the common dolphin, although the above discussion of the low power of sex-specific F ST and assignment methods should be noted.
Theoretical models suggest that, in promiscuous mating systems, female-biased dispersal is likely to develop if local resource competition affects female reproductive success (Perrin and Mazalov 2000). When common dolphins hunt, the process of aggregation of fish schools requires cooperation between individuals. Once the fish is trapped against the surface, individual dolphins might then compete for access to the disoriented fish. Studies on common dolphin feeding ecology show that females and juveniles often exhibit different feeding strategies to adult males (Silva 1999; Nino-Torres et al. 2006). In a scenario of competitive access to food, juveniles and females can be at a disadvantage, as their smaller size makes them less able to compete (Ruckstuhl and Neuhaus 2000; Meynier et al. 2008).
In these circumstances, during periods of reduced prey abundance, females could have reduced access to prey due to increased competition with larger males, particularly if the female is pregnant or accompanied by a calf. Therefore, females benefit the most from dispersing elsewhere, where prey may be more abundant and local resource competition is less intense. The only study so far available on common dolphin dispersing behaviour reports the long-distance dispersal of a female dolphin accompanied by a calf, which was followed by site fidelity to new area for at least 1 year (Genov et al. 2012). Long-distance dispersal means here that the animals do not expand their natural range continuously, but ignore some close-by locations with suitable habitats and settle farther apart.
A calf accompanying a dispersing female could be a male, in which case male dispersal would also occur. Given a sex ratio of 1:1, this would effectively skew dispersal towards females, as only half of the dispersal events would statistically be expected to involve a male. This would be consistent with the idea discussed above that sex dispersal asymmetry, if present, is likely to be subtle in this system.
Kinship patterns and large-scale panmixia
It thus appears that the fluid social structure in short-beaked common dolphins and the genetic panmixia over large spatial scales in the Northeast Atlantic and the Mediterranean Sea may result from their specific feeding ecology. Common dolphins feed mainly on high-energy epipelagic schooling fish (Spitz et al. 2010), through a technique that requires cooperation between large numbers of individuals. This technique thus promotes social interactions between individuals irrespective of their level of relatedness. Distribution of such prey items tends to be seasonal and is dependent on specific environmental characteristics, making its exact distribution variable in time. In the Portuguese coast, common dolphin’s preferred prey is the sardine (Sardina pilchardus; Silva 1999), whose patterns of abundance and distribution are known to fluctuate between years (Santos 2001; Borges et al. 2003). Some regions might have more favourable conditions, and therefore, the appearance of suitable prey can be predictable to some extent. This increases the costs of exploring larger unfamiliar areas where prey abundance patterns are uncertain. This can lead to habitat dependence and natal philopatry, but also promote the long-distance dispersal behaviour when local prey resources become depleted.
Conclusions
The results of this study show that common dolphin (D. delphis) large-scale panmixia in Europe (Moura et al. 2013) likely results from long-distance dispersal of individuals questing for seasonal prey, rather than from extreme promiscuity and fluid social structure. Our results provide new insight into the social and mating systems of this poorly studied species, for which field observation data on mating behaviour are scarce. Our analysis of the temporal and geographic distribution of kin groups provided support for a fission-fusion social structure in common dolphins. Furthermore, this analysis indicates female-biased dispersal, which had so far remained undetected. This dispersal pattern is consistent with the occurrence of competition for an unpredictable prey resource and the lack of paternal care in this species. Female-biased dispersal is uncommon in mammals, and therefore, more research based on a larger sample size is needed to confirm this pattern. Despite the limitations resulting from relatively small sample sizes, our approach demonstrates the inference potential to be gained from detailed kinship studies, and highlights the importance of collecting long-term genetic data in gaining a better understanding of the ecology of elusive long-lived animals.
References
Amaral AR, Beheregaray LB, Bilgmann K, Boutov D, Freitas L, Robertson KM, Sequeira M, Stockin KA, Coelho M, Möller LM (2012) Seascape genetics of a globally distributed, highly mobile marine mammal: the short-beaked common dolphin (genus Delphinus). PLoS One 7:e31482
Amos B (1999) Technical comment: cultural and genetic evolution in whales. Science 284:2055
Bérubé M, Palsbøll P (1996) Identification of sex in cetaceans by multiplexing with three ZFX and ZFY specific primers. Mol Ecol 5:283–287
Bilgmann K, Möller LM, Harcourt RG, Gales R, Beheregaray LB (2008) Common dolphins subject to fisheries impacts in Southern Australia are genetically differentiated: implications for conservation. Anim Conserv 11:518–528
Bilgmann K, Guido PJ, Beheregaray LB, Zanardo N, Möller LM (2014) Multiple management units of short-beaked common dolphins subject to fisheries bycatch off southern and southeastern Australia. Mar Ecol Prog Ser 500:265–279
Blouin MS, Parsons M, Lacaille V, Lotz S (1996) Use of microsatellite loci to classify individuals by relatedness. Mol Ecol 5:393–401
Borges M, Santos A, Crato N, Mendes H, Mota B (2003) Sardine regime shifts off Portugal: a time series analysis of catches and wind conditions. Sci Mar 67:235–244
Bradley BJ, Doran-Sheehy DM, Vigilant L (2007) Potential for female kin associations in wild western gorillas despite female dispersal. Proc R Soc Lond B 274:2179–2185
Bruno S, Politi E, Bearzi G (2004) Social organization of a common dolphin community in the eastern Ionian Sea: evidence of a fluid fission-fusion society. Eur Res Cetac 15:49–51
Catchpole EA, Fan Y, Morgan BJT, Clutton-Brock TH, Coulson T (2004) Sexual dimorphism, survival and dispersal in red deer. J Agric Biol Environ Stat 9:1–26
Clutton-Brock TH (1989) Female transfer and inbreeding avoidance in social mammals. Nature 337:70–72
Clutton-Brock TH, Lukas D (2012) The evolution of social philopatry and dispersal in female mammals. Mol Ecol 21:472–492
Connor RC (2000) Group living in whales and dolphins. In: Mann J, Connor RC, Tyack PL, Whitehead H (eds) Cetacean societies: field studies of dolphins and whales. University of Chicago Press, Chicago, pp 199–218
Connor RC, Krützen M (2015) Male dolphin alliances in Shark Bay: changing perspectives in a 30-year study. Anim Behav 103:223–235
Connor RC, Smolker RA, Richards AF (1992) Two levels of alliance formation among male bottlenose dolphins (Tursiops sp.) Proc Natl Acad Sci USA 89:987–990
Coscarella MA, Gowans S, Pedraza SN, Crespo EA (2011) Influence of body size and ranging patterns on delphinid sociality: associations among Commerson’s dolphins. J Mammal 92:544–551
Díaz-Muñoz SL, DuVal EH, Krakauer AH, Lacey EA (2014) Cooperating to compete: altruism, sexual selection and causes of male reproductive cooperation. Anim Behav 88:67–78
Dobson FS (1982) Competition for mates and predominant juvenile male dispersal in mammals. Anim Behav 30:1183–1192
Ford JKB, Ellis GM, Balcomb KC (2000) Killer whales: the natural history and genealogy of Orcinus orca in British Columbia and Washington state. UBC Press, Vancouver
Genov T, Bearzi G, Bonizzoni S, Tempesta M (2012) Long-distance movement of a lone short-beaked common dolphin Delphinus delphis in the central Mediterranean Sea. Mar Biodivers Rec 5:e9
Goudet J (2001) FSTAT, a program to estimate and test gene diversities and fixation indices, version 2.9.3, http://www.unil.ch/izea/softwares/fstat.html
Goudet J, Perrin N, Waser P (2002) Tests for sex-biased dispersal using bi-parentally inherited genetic markers. Mol Ecol 11:1103–1114
Gowans S, Würsig B, Karczmarski L (2007) The social structure and strategies of delphinids: predictions based on an ecological framework. Adv Mar Biol 53:195–294
Greenwood PJ (1980) Mating systems, philopatry and dispersal in birds and mammals. Anim Behav 28:1140–1162
Hammer Ø, Harper DAT, Ryan PD (2001) PAST: Paleontological Statistics Software Package for education and data analysis. Palaeontol Electron 4:1–9
Heimlich-Boran JR (1993) Social organisation of the short-finned pilot whale, Globicephala macrorhynchus, with special reference to the comparative social ecology of delphinids. PhD thesis, University of Cambridge
Kalinowski ST, Taper M, Marshall TC (2007) Revising how the computer program cervus accommodates genotyping error increases success in paternity assignment. Mol Ecol 16:1099–1106
Konovalov DA, Manning C, Henshaw MT (2004) KINGROUP: a program for pedigree relationship reconstruction and kin group assignments using genetic markers. Mol Ecol Notes 4:779–782
Lawson Handley LJ, Perrin N (2007) Advances in our understanding of mammalian sex-biased dispersal. Mol Ecol 16:1559–1578
Lyrholm T, Gyllensten U (1998) Global matrilineal population structure in sperm whales as indicated by mitochondrial DNA sequences. Proc R Soc Lond B 265:1679–1684
MacLeod CD (2010) The relationship between body mass and relative investment in testes mass in cetaceans: implications for inferring interspecific variations in the extent of sperm competition. Mar Mamm Sci 26:370–380
Mason S, Salgado Kent C, Donnelly D, Weir J, Bilgmann K (2016) Atypical residency of short-beaked common dolphins (Delphinus delphis) to a shallow, urbanised embayment in south-eastern Australia. R Soc Open Sci 3:160478
Meynier L, Pusineri C, Spitz J, Santos MB, Pierce GJ, Ridoux V (2008) Intraspecific dietary variation in the short-beaked common dolphin Delphinus delphis in the Bay of Biscay: importance of fat fish. Mar Ecol Prog Ser 354:277–287
Mirimin L, Westgate A, Rogan E, Rosel P, Read A, Coughlan J, Cross T (2009) Population structure of short-beaked common dolphins (Delphinus delphis) in the North Atlantic Ocean as revealed by mitochondrial and nuclear genetic markers. Mar Biol 156:821–834
Möller LM (2012) Sociogenetic structure, kin associations and bonding in delphinids. Mol Ecol 21:745–764
Möller LM, Beheregaray LB, Harcourt RG, Krützen M (2001) Alliance membership and kinship in wild male bottlenose dolphins (Tursiops aduncus) of southeastern Australia. Proc R Soc Lond B 268:1941–1947
Möller LM, Beheregaray LB, Allen SJ, Harcourt RG (2006) Association patterns and kinship in female Indo-Pacific bottlenose dolphins (Tursiops aduncus) of southeastern Australia. Behav Ecol Sociobiol 61:109–117
Möller LM, Pedoni F, Allen S, Bilgmann K, Corrigan S, Beheregaray LB (2011) Fine-scale genetic structure in short-beaked common dolphins (Delphinus delphis) along the East Australian Current. Mar Biol 158:113–126
Moura AE, Natoli A, Rogan E, Hoelzel AR (2013) Atypical panmixia in a European dolphin species (Delphinus delphis): implications for the evolution of diversity across oceanic boundaries. J Evol Biol 26:63–75
Moura AE, Tsingarska E, Dąbrowski MJ, Czarnomska SD, Jędrzejewska B, Pilot M (2014) Unregulated hunting and genetic recovery from a severe population decline: the cautionary case of Bulgarian wolves. Conserv Genet 15:405–417
Murphy S, Collet A, Rogan E (2005) Mating strategy in the male common dolphin (Delphinus delphis): what gonadal analysis tells us. J Mammal 86:1247–1258
Musiani M, Leonard JA, Cluff HD, Gates CC, Mariani S, Paquet PC, Vilà C, Wayne RK (2007) Differentiation of tundra/taiga and boreal coniferous forest wolves: genetics, coat colour and association with migratory caribou. Mol Ecol 16:4149–4170
Nagy M, Heckel G, Voigt CC, Mayer F (2007) Female-biased dispersal and patrilocal kin groups in a mammal with resource-defence polygyny. Proc R Soc Lond B 274:3019–3025
Natoli A, Cañadas A, Peddemors VM, Aguilar A, Vaquero C, Fernández-Piqueras P, Hoelzel AR (2006) Phylogeography and alpha taxonomy of the common dolphin (Delphinus sp.) J Evol Biol 19:943–954
Natoli A, Cañadas A, Vaquero C, Politi E, Fernandez-Navarro P, Hoelzel AR (2008) Conservation genetics of the short-beaked common dolphin (Delphinus delphis) in the Mediterranean Sea and in the eastern North Atlantic Ocean. Conserv Genet 9:1479–1487
Nino-Torres CA, Gallo-Reynoso JP, Galvan-Magana F, Escobar-Briones E, Macko SA (2006) Isotopic analysis of ∆13C, ∆15N, and ∆34S: a feeding tale in teeth of the longbeaked common dolphin, Delphinus capensis. Mar Mamm Sci 22:831–846
Oremus M, Gales R, Kettles H, Baker CS (2013) Genetic evidence of multiple matrilines and spatial disruption of kinship bonds in mass strandings of long-finned pilot whales, Globicephala melas. J Hered 104:301–311
Parsons KM, Durban JW, Claridge DE, Balcomb KC, Noble LR, Thompson PM (2003) Kinship as a basis for alliance formation between male bottlenose dolphins, Tursiops truncatus, in the Bahamas. Anim Behav 66:185–194
Peakall ROD, Smouse PE (2006) Genalex 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Pérez-González J, Carranza J (2009) Female-biased dispersal under conditions of low male mating competition in a polygynous mammal. Mol Ecol 18:4617–4630
Perrin W (2009) Common dolphin. In: Perrin W, Würsig B (eds) Encyclopedia of marine mammals. Academic Press, New York
Perrin N, Mazalov V (1999) Dispersal and inbreeding avoidance. Am Nat 154:282–292
Perrin N, Mazalov V (2000) Local competition, inbreeding, and the evolution of sex-biased dispersal. Am Nat 155:116–127
Pilot M, Jędrzejewski W, Branicki W, Sidorovich VE, Jędrzejewska B, Stachura K, Funk SM (2006) Ecological factors influence population genetic structure of European grey wolves. Mol Ecol 15:533–4553
Pilot M, Dąbrowski MJ, Jancewicz E, Schtickzelle N, Gliwicz J (2010a) Temporally stable genetic variability and dynamic kinship structure in a fluctuating population of the root vole Microtus oeconomus. Mol Ecol 19:2800–2812
Pilot M, Dahlheim ME, Hoelzel AR (2010b) Social cohesion among kin, gene flow without dispersal and the evolution of population genetic structure in the killer whale (Orcinus orca). J Evol Biol 23:20–31
Ruckstuhl K, Neuhaus P (2000) Sexual segregation in ungulates: a new approach. Behaviour 137:361–377
Santos A (2001) Sardine and horse mackerel recruitment and upwelling off Portugal. ICES J Mar Sci 58:589–596
Santos MCO, Rosso S (2008) Social organization of marine Tucuxi dolphins, Sotalia guianensis, in the Cananéia Estuary of southeastern Brazil. J Mammal 89:347–355
Silva MA (1999) Diet of common dolphins, Delphinus delphis, off the Portuguese continental coast. J Mar Biol Assoc UK 79:531–540
Spitz J, Mourocq E, Leauté J-P, Quéro J-C, Ridoux V (2010) Prey selection by the common dolphin: fulfilling high energy requirements with high quality food. J Exp Mar Biol Ecol 390:73–77
Sweitzer RA, Berger J (1998) Evidence for female-biased dispersal in North American porcupines (Erethizon dorsatum). J Zool 244:159–166
Van Horn RC, Altmann J, Alberts SC (2008) Can’t get there from here: inferring kinship from pairwise genetic relatedness. Anim Behav 75:1173–1180
Vigilant L, Guschanski K (2009) Using genetics to understand the dynamics of wild primate populations. Primates 50:105–120
Viricel A, Strand AE, Rosel PE, Ridoux V, Garcia P (2008) Insights on common dolphin (Delphinus delphis) social organization from genetic analysis of a mass-stranded pod. Behav Ecol Sociobiol 63:173–185
Watts DP (1998) Coalitionary mate guarding by male chimpanzees at Ngogo, Kibale National Park, Uganda. Behav Ecol Sociobiol 44:43–55
Wiszniewski J, Beheregaray LB, Allen SJ, Möller LM (2010) Environmental and social influences on the genetic structure of bottlenose dolphins (Tursiops aduncus) in southeastern Australia. Conserv Genet 11:1405–1419
Zanardo N, Bilgmann K, Parra GJ, Möller LM (2016) Socio-genetic structure of short-beaked common dolphins in southern Australia. J Zool 299:89–97
Acknowledgments
The authors would like to acknowledge the funding provided by the Portuguese Fundação para a Ciência e Tecnologia (PhD grant SFRH/BD/28012/2006) and the logistical support provided by Marina de Portimão, AngelPilot and Nautiradar. The authors also acknowledge all the crew of ‘Clavadel’ for their tireless effort and dedication in the collection of biopsy samples. We also acknowledge the invaluable contribution of three anonymous reviewers, which greatly improved the manuscript.
Author information
Authors and Affiliations
Contributions
LB carried out statistical analyses, result interpretation and contributed to writing the manuscript. KS contributed to result interpretation, critically revised it and wrote the manuscript. MP designed the study, critically revised the result interpretation and manuscript and contributed to the biopsy collection. AEM designed the study, collected all biopsy samples, produced microsatellite genotypes and revised the manuscript. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Funding
Sample collection was supported through logistic support from the companies Marina de Portimão, AngelPilot and Nautiradar. Lab work was funded through a PhD grant (SFRH/BD/28012/2006) awarded to AEM by the Portuguese Fundação para a Ciência e Tecnologia.
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All applicable international, national and/or institutional guidelines for the care and the use of animals were followed. We only sampled adults and biopsy samples were collected under the permit from the Portuguese Institute for the Conservation of Nature (ICN).
Informed consent
The research presented in this manuscript did not involve human participants.
Data availability
The datasets analysed during the current study are available in the Dryad repository, http://datadryad.org/resource/doi:10.5061/dryad.15nk1.
Additional information
Communicated by L. M. Moller
Electronic supplementary material
ESM 1
(DOCX 60 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Ball, L., Shreves, K., Pilot, M. et al. Temporal and geographic patterns of kinship structure in common dolphins (Delphinus delphis) suggest site fidelity and female-biased long-distance dispersal. Behav Ecol Sociobiol 71, 123 (2017). https://doi.org/10.1007/s00265-017-2351-z
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00265-017-2351-z