Abstract
Purpose
Immune checkpoint inhibitors have revolutionized the treatment of renal cell carcinoma (RCC), but many patients do not respond to therapy and the majority develop resistant disease over time. Thus, there is increasing need for alternative immunomodulating agents. The co-inhibitory molecule T-cell immunoglobulin and ITIM domain (TIGIT) may play a role in resistance to approved immune checkpoint inhibitors and is being investigated as a potential therapeutic target. The purpose of this study was to quantify TIGIT positivity in tumor-infiltrating T cells in RCC.
Methods
We employed tissue microarrays containing specimens from primary RCC tumors, adjacent normal renal tissue, and RCC metastases to quantify TIGIT within tumor-infiltrating CD3+ T cells using quantitative immunofluorescent analysis. We also compared these results to TIGIT+ CD3+ levels in four other tumor types (melanoma, non-small cell lung, cervical, and head and neck cancers).
Results
We did not observe significant differences in TIGIT positivity between primary RCC tumors and patient-matched metastatic samples. We found that the degree of TIGIT positivity in RCC is comparable to that in lung cancer but lower than that in melanoma, cervical, and head and neck cancers. Correlation analysis comparing TIGIT positivity to previously published, patient-matched spatial proteomic data by our group revealed a negative association between TIGIT and the checkpoint proteins PD-1 and LAG3.
Conclusion
Our findings support careful evaluation of TIGIT expression on T cells in primary or metastatic RCC specimens for patients who may be treated with TIGIT-targeting antibodies, as increased TIGIT positivity might be associated with a greater likelihood of response to therapy.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In renal cell carcinoma (RCC), immune checkpoint inhibitor (ICI)-based regimens have significantly improved overall survival compared to previous standard-of-care tyrosine kinase inhibitors (TKIs). For example, a small minority of patients treated with the combination of ipilimumab (anti-CTLA-4) and nivolumab (anti-PD-1) remain in complete response and off therapy after more than 6 years [1, 2]. However, resistance to ICI-based regimens remains a challenge, highlighting a need for alternative immunotherapeutic strategies.
Other co-inhibitory receptors expressed on immune cells, such as LAG3, TIM-3 and TIGIT, are promising targets [3]. Expression of these co-inhibitory receptors results in increased tumor tolerance and T cell anergy through pathways parallel to the canonical PD-1/PD-L1 and CTLA-4/CD80/86 axes. Engagement of TIGIT on T cells downregulates T cell receptor complex components, impairing recognition of tumor antigens [4]. TIGIT is highly expressed on immunosuppressive regulatory T cells (Tregs) and natural killer (NK) cells, stimulating production of the anti-inflammatory cytokine IL-10 and downregulating tumor surveillance [3, 5]. Previous work has suggested a TIGIT receptor-positive cell density of 0.4% in RCC primary tumors and 2.1% in metastases, notably lower than receptor-positive densities for LAG3 and TIM-3; expression of these co-inhibitory receptors was also largely mutually exclusive and could define distinct tumor subtypes [6]. Single-cell RNA-sequencing studies in RCC have further defined TIGIT expression patterns in the tumor microenvironment (TME). One such study demonstrated enrichment of TIGIT, as well as other immune checkpoints, on terminally exhausted CD8+ T cells in the primary tumors of patients that would go on to develop more advanced disease [7]. TIGIT’s role as part of a posited dysfunctional immune circuit involving T cells and macrophages leading to more advanced disease in RCC was further confirmed in this study through flow cytometry analysis of TIGIT expression on terminally exhausted CD8+ T cells paired with expression of CD155, a TIGIT ligand, on M2-like, immunosuppressive macrophages. TIGIT expression thus may serve as a biomarker to predict resistance to immunotherapy or as a therapeutic target itself.
TIGIT blockade is being evaluated in multiple clinical trials, including phase III studies for advanced non-small cell lung cancer (NSCLC) and melanoma, and phase II basket trials for solid tumors, although none treating RCC specifically (NCT04294810, NCT05665595, NCT04693234, NCT03708224). Of note, only patients with elevated PD-L1 expression were found to benefit from TIGIT blockade in one phase II trial [8]. The role of TIGIT expression on CD3+ T cells as a predictive biomarker is unknown.
Given the interest in anti-TIGIT therapies in solid tumors, our purpose was to evaluate the degree of TIGIT expression on tumor-infiltrating T cells in RCC at different anatomic sites and relative to other cancer types. We performed immunofluorescent analysis of TIGIT positivity on CD3+ T cells on cohorts of primary RCC tumors and matched adjacent normal kidney samples, matched RCC primary and metastatic tumors, and cohorts from four other tumor types (melanoma, NSCLC, cervical, and head and neck cancer). Overall, we found no differences between RCC primary tumors and metastases and TIGIT positivity in RCC was comparable to NSCLC but lower than in the other cancer types.
Materials and methods
Tissue microarrays
We performed these studies using two previously reported RCC tissue microarrays (TMAs) (Table 1) [9,10,11,12,13]. YTMA84-A1 and YTMA84-B1 contained primary tumor tissue and adjacent normal renal parenchyma, and YTMA166-1 contained paired primary tumors and metastases. The TMAs consisted of 0.6 mm cores spaced 0.8 mm apart. Our positive control YTMA462 contained lymphoid tissue from tonsil and spleen (n = 11). YTMA226-5 was used for lung cancer (n = 22), YTMA383-1 for cervical cancer (n = 23), YTMA335-8 for melanoma (n = 13), and YTMA275 for head and neck cancer (n = 27).
Immunostaining, multispectral image acquisition and analysis
Briefly [14], mouse anti-CD3 antibody (Invitrogen MA5-12577) was diluted 1:100 and incubated overnight at 4 °C. Signal amplification was performed using anti-mouse EnVision antibody-HRP (Dako K4001) and HRP-activated-Cy5-tyramide (1:50; Akoya Biosciences) following the manufacturer’s protocol. HRP-quenching was done with 100 mM benzoic hydrazide + 50 mM hydrogen peroxide. Slides were incubated for 2 h at room temperature with anti-TIGIT antibody (Abcam 243903) diluted 1:400. Anti-TIGIT signal was amplified using anti-rabbit EnVision system-HRP (Dako K4003) and Cy3-Tyramide (1:50; Akoya Biosciences). Slides were incubated with 4,6-diamidino-2-phenylindole (DAPI) and mounted with ProLong mounting medium (ProLong Gold; Invitrogen, Carlsbad, CA, USA).
Monochromatic image acquisition was performed using Aquasition (HistoRx) software on images captured through a microscopy-based multiplex imaging device and microarray reader (PM-3000; HistoRx). Due to generally low levels of TIGIT positivity on CD3+ cells on the RCC TMAs, cell counts were manually quantified. On slides with high density of CD3 staining, cell counts were extrapolated from representative quadrants. For this study, Cy3+, Cy5− cells were excluded from analysis.
Statistics
Statistical analysis and graphing were performed using GraphPad Prism 10 (GraphPad Software, Inc.). TCGA data were taken from cBioPortal [15,16,17]. Some tumor samples on YTMA166 were represented by multiple biologic replicates; when present, replicate CD3 counts and levels of TIGIT positivity were averaged. Kruskal–Wallis test with Dunn’s test for multiple comparisons was used to compare TIGIT positivity levels between tumor types. Mann–Whitney tests were used for testing mean differences between samples. For matched-pair analysis, Wilcoxon matched-pair test was used. ANOVA was used to compare TIGIT+ T cells across metastatic sites. Chi-squared test was used to test differences in TIGIT positivity across tumor types. Correlation analyses were performed using Spearman’s rank test; outliers were detected and removed with formal outlier testing using Grubb’s test with a P < 0.01 considered significant. All other statistical testing was performed with a two-sided P < 0.05 considered significant.
Results
High TIGIT expression in RCC is associated with increased tumor grade, stage, and decreased survival
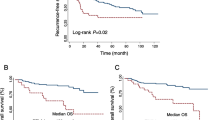
We used a single-cell transcriptomic dataset to confirm the expression of TIGIT and its ligands, NECTIN2 and PVR, in the RCC TME [18]. We found that TIGIT is mainly expressed on T cells, while the TIGIT ligands, primarily NECTIN2, are expressed on tumor-associated macrophages (TAMs), tumor cells, and endothelial cells, in line with findings from other RCC single-cell RNA studies (Fig. 1A) [7]. We then queried publicly available TCGA PanCancer genomic datasets on cBioPortal to compare TIGIT mRNA expression in RCC with other solid malignancies [15,16,17]. We found that expression of TIGIT was relatively high in clear cell RCC (ccRCC), but lower than in other malignancies, including melanoma, NSCLC, head and neck, and cervical cancer (Supplementary (Supp.) Fig. S1A). With prior studies demonstrating a relationship between TIGIT expression and the development of more advanced disease at the single-cell level but with a relatively limited number of patient samples [7], we next sought to determine the relationship between TIGIT expression and overall survival (OS) in ccRCC using the larger TCGA PanCancer dataset. We categorized patients as having either high, intermediate, or low TIGIT expression using z-scores [15, 19]. Z-scores > 1 were defined as “high” and < − 1 as “low.” Comparing high to low TIGIT expressors, high TIGIT expression was associated with worse 5-year OS (HR = 2.05, p = 0.015) (Fig. 1B). Advanced histologic grade and stage were also associated with higher TIGIT levels, in accord with the single-cell transcriptomic and flow cytometry findings from the prior study (Fig. 1C, D) [7]. However, a Cox proportional hazards model incorporating TIGIT levels, grade, and stage showed that TIGIT expression may not be an independent prognostic factor (Supp. Table S1).
A Dot plot of single-cell mRNA expression for TIGIT, PVR, and NECTIN2 in the RCC TME, by cell type, from the Broad Institute’s Single Cell portal and [18]. B Kaplan–Meier curves of overall survival for ccRCC patients based on level of TIGIT mRNA expression, with high TIGIT expressors having a z-score > 1, and low TIGIT expressors having a z-score < − 1. Expression and survival data from the TCGA PanCancer Atlas on cBioPortal. TIGIT mRNA expression in ccRCC by C histologic grade and D stage. Statistical testing was performed for C, D using ANOVA Dunnett test for multiple comparisons, with Grade or Stage I as the control. *p < 0.05; **p < 0.01;***p < 0.001; ****p < 0.0001
TIGIT positivity is similar between RCC primary tumors and metastases
We quantified the percentage of T cells (CD3+) that were TIGIT-positive in RCC through immunofluorescence. As TIGIT is highly expressed in tonsillar tissue, we employed a TMA of normal tonsil as a positive staining control and to establish the staining conditions (Fig. 2A) [20]. An example of TIGIT-positive staining on T cells in an RCC TMA is shown in Fig. 2B.
A Exemplary immunofluorescent staining of TIGIT and CD3 in a tonsillar tissue specimen, which served as a positive control. B Representative stain of TIGIT and CD3 in RCC primary tumor tissue. Percentage of CD3 + cells expressing TIGIT in: C unmatched and D matched adjacent normal renal tissue and primary RCC tumors from YTMA84; and E unmatched and F matched RCC primary tumors and metastases from YTMA166; and G multiple anatomic sites of RCC metastases from YTMA166. H) Absolute CD3 + cell counts in adjacent normal renal tissue, RCC primary tumors, and metastases, with mean and 95% CI graphed. I Distribution of high (≥ 5%), low (< 5%), and absent TIGIT positivity levels comparing unmatched primary RCC tumors versus metastases from YTMA166. Statistical tests performed with T test or ANOVA. *p < 0.05; **p < 0.01; ***p < 0.001
We compared the percentage of TIGIT-expressing T cells between primary RCC tumors, adjacent normal tissue, and metastatic sites. There were no differences between normal adjacent renal tissue (n = 54) and RCC primary tumors (n = 191) in unmatched and matched-pair (n = 20) patient samples (Fig. 2C, D). Likewise, the density of TIGIT+ T cells was similar between primary (n = 36) and metastatic (n = 36) tumor tissue in unmatched as well as matched-pair (n = 15) analysis (Fig. 2E, F). TIGIT positivity was also not significantly different at various RCC anatomic sites of metastasis (Fig. 2G). Among clinical factors, we found a significant but weak correlation with tumor size (R2 = 0.062, p = 0.006) (Supp. Fig. S2). Additionally, RCC metastases had significantly higher levels of T cell infiltration compared to normal renal parenchyma, but not relative to primary tumor specimens (Fig. 2H). RCC primary tumors and metastases had similar percentages of TIGIT-negative samples (p = 0.805) (Fig. 2I). Among the TIGIT+ samples (TIGIT+ T cells > 0%), we further dichotomized based on degree of TIGIT T cell positivity, defining high expressors as having ≥ 5% positive T cells and low expressors as having < 5%. Still, the proportion of high expressors was comparable in primary tumors and metastases (p = 0.332).
TIGIT positivity on tumor-infiltrating T cells is comparable between RCC and NSCLC, but higher in other tumor types
Given the high levels of TIGIT mRNA expression in melanoma, NSCLC, head and neck, and cervical cancer, we also stained TMAs containing these tumor types for CD3 and TIGIT. The percentage of T cells expressing TIGIT was significantly higher in melanoma, cervical, and head and neck cancer relative to RCC primary tumors (Fig. 3A). While the measurement of TIGIT positivity is intrinsically normalized to T cell density, we additionally quantified absolute T cell counts across tumor types. We observed lower degrees of T cell infiltration in RCC compared to NSCLC and cervical cancer, which had the highest T cell counts among the tumor types evaluated (Fig. 3B). Across RCC primary tumors, 39.6% of samples had any level of TIGIT+ T cells (i.e., TIGIT+ T cells > 0%) (Fig. 3C). Melanoma and cervical cancer had the highest proportion of TIGIT+ tumors at > 60%, although only cervical cancer had a significantly higher degree of positivity compared to RCC (p = 0.0491).
A Violin plots showing percentage of TIGIT + CD3 cells across multiple tumor types. B Absolute CD3 + cell counts across RCC primary tumor samples and the other indicated cancer types. C Percentage of TIGIT + (> 0%) samples in each of the indicated cancer types. RCC bar includes primary tumors from YTMA84 and YTMA166. Significance tested by Chi-squared test. *p < 0.05; **p < 0.01; ***p < 0.001, ****p < 0.0001
TIGIT positivity is negatively correlated with PD-1 and LAG3 protein expression in RCC
We previously performed spatial proteomic profiling on overlapping samples from the RCC TMAs to characterize expression patterns of multiple tumor immunomarkers, including CD3, PD-1, PD-L1, LAG3, TIM-3, and CTLA-4 in immune cells (CD45+), and PD-L1 in tumor cells (CK+) [9, 10]. To better understand the relationship between TIGIT and these other immune proteins, we performed Spearman tests for correlation between the degree of TIGIT positivity and these markers, looking separately at primary tumors, normal adjacent renal tissue, and metastases. Our manual CD3 counts correlated with the degree of CD3 expression as determined by the digital spatial profiling method (p = 0.019, Supp. Fig S3). There were no significant correlations between TIGIT+ T cell counts and these markers in primary tumors and normal renal tissue (Supp. Fig. S4A, B). Among metastases only, there was a negative correlation with PD-1 expression (r = −0.524, p = 0.048), and among all tumor samples (primary tumors and metastases), there were negative correlations between TIGIT and PD-1 (r = −0.301, p = 0.032), and TIGIT and LAG3 (r = −0.394, p = 0.005) (Fig. 4A, B and Supp. Fig S4C, D).
Percentage of TIGIT + CD3 cells graphed against patient-matched spatial proteomic profiling of the indicated immune cell (CD45 +) markers in A RCC metastases, and B all RCC tumor samples. Spearman rank test was used to assess the correlation between percentage of TIGIT + CD3 cells and each immune marker. Best fit linear regression lines are shown
Discussion
Our analysis aimed to better characterize expression patterns of the immune checkpoint protein TIGIT on tumor-infiltrating T cells in RCC. Using TCGA PanCancer transcriptomic data, we found that TIGIT mRNA levels are relatively high in ccRCC compared to most other tumor types, and that TIGIT expression was positively correlated with higher histologic grade and stage, in accord with prior single-cell transcriptomic findings. Immunofluorescent studies showed that TIGIT was detectable on tumor-infiltrating T cells in ~ 40% of RCC patient samples, although only a small subset had ≥ 5% TIGIT-positive T cells. While the degree of TIGIT positivity was significantly higher in melanoma, head and neck, and cervical cancer compared to RCC, there was no significant difference in TIGIT+ T cell infiltration between RCC and NSCLC, a tumor type where anti-TIGIT therapies are being extensively studied with numerous late-stage clinical trials underway [21].
Additionally, we found no significant differences between primary RCC tumors and metastases, and between different metastatic sites. We note that many patients with metastatic RCC might only have previously resected large primary tumor samples available for tissue analysis. Our data suggest that sampling from either primary or metastatic sites may be sufficient to determine TIGIT T cell positivity.
Ongoing clinical trials are investigating anti-TIGIT agents in combination with other ICIs based partially on the hypothesis that upregulation of TIGIT may serve as a mechanism of resistance to first-line ICIs. We found a negative correlation between TIGIT positivity on T cells and PD-1 and LAG3 protein levels on tumor-infiltrating immune cells, supporting the hypothesis that TIGIT upregulation may compensate for low expression levels of other immune checkpoint proteins in RCC. These findings are also in agreement with previous research noting the presence of phenotypically distinct tumor clusters, where TIM-3, LAG3 and TIGIT expression in the RCC TME appear to be mutually exclusive [6]. This study further showed that each cluster may be associated with distinct genomic alterations, notably p53/cell cycle alterations in LAG3-positive tumors. Further studies, such as single-cell spatial multi-omic investigation of patient samples pre- and post-ICIs, are needed to help elucidate the molecular drivers regulating the expression of specific co-inhibitory receptors in the RCC TME.
One limitation of our study is that we focused on TIGIT expression on CD3+ T cells, while there are other immune populations that express this protein, such as NK cells. Future studies should focus on determining the expression of TIGIT in other immune cell types in RCC, including NK cells, and aim to further characterize expression in T cell subsets, including cytotoxic CD8+, CD4+, and Treg cells. We also note that the percentage of PD-L1 expressing cells in melanoma and RCC is low compared to lung cancer, yet response rates to PD-1/L1 inhibitors are higher in melanoma and RCC [22]. Similar to PD-L1 positivity as a predictive biomarker for PD-1/L1 inhibitors, TIGIT positivity might not correlate with response to anti-TIGIT antibodies.
In summary, we quantified the percentage of TIGIT-positive T cells in RCC tumors across different anatomic sites of disease and compared to other tumor types. We detected some level of TIGIT positivity in ~ 40% of RCC specimens, comparable to NSCLC, but lower than melanoma, head and neck, and cervical cancer cohorts. Additionally, the percentage of T cells expressing TIGIT in RCC metastatic samples was similar to primary tumors, indicating that sampling from either site is sufficient to determine TIGIT T cell positivity. As ongoing clinical trials investigating anti-TIGIT agents in other malignancies proceed, the results of this study suggest that TIGIT blockade may need to be selectively employed in patients with RCC.
Data availability
The dataset upon which this study is based is available upon reasonable request.
References
Motzer RJ et al (2018) Nivolumab plus Ipilimumab versus sunitinib in advanced renal-cell carcinoma. N Engl J Med 378(14):1277–1290
Tannir NM et al (2024) Nivolumab plus ipilimumab (NIVO+IPI) vs sunitinib (SUN) for first-line treatment of advanced renal cell carcinoma (aRCC): Long-term follow-up data from the phase 3 checkmate 214 trial. J Clin Oncol 42(4_suppl):363–363
Anderson AC, Joller N, Kuchroo VK (2016) Lag-3, Tim-3, and TIGIT: co-inhibitory receptors with specialized functions in immune regulation. Immunity 44(5):989–1004
Joller N et al (2011) Cutting edge: TIGIT has T Cell-intrinsic inhibitory functions. J Immunol 186(3):1338
Chauvin JM, Zarour HM (2020) TIGIT in cancer immunotherapy. J Immunother Cancer 8(2):e000957
Takamatsu K et al (2021) Profiling the inhibitory receptors LAG-3, TIM-3, and TIGIT in renal cell carcinoma reveals malignancy. Nat Commun 12(1):5547
Braun DA et al (2021) Progressive immune dysfunction with advancing disease stage in renal cell carcinoma. Cancer Cell 39(5):632-648.e8
Cho BC et al (2022) Tiragolumab plus atezolizumab versus placebo plus atezolizumab as a first-line treatment for PD-L1-selected non-small-cell lung cancer (CITYSCAPE): primary and follow-up analyses of a randomised, double-blind, phase 2 study. Lancet Oncol 23(6):781–792
Schoenfeld DA et al (2022) Location matters: LAG3 levels are lower in renal cell carcinoma metastatic sites compared to primary tumors, and expression at metastatic sites only may have prognostic importance. Front Oncol 12:990367
Schoenfeld DA et al (2023) Immune dysfunction revealed by digital spatial profiling of immuno-oncology markers in progressive stages of renal cell carcinoma and in brain metastases. J Immunother Cancer 11(8):e007240
Xie Z et al (2016) MET Inhibition in Clear Cell Renal Cell Carcinoma. J Cancer 7(10):1205–1214
Baine MK et al (2015) Characterization of tumor infiltrating lymphocytes in paired primary and metastatic renal cell carcinoma specimens. Oncotarget 6(28):24990–25002
Barr ML et al (2015) PAX-8 expression in renal tumours and distant sites: a useful marker of primary and metastatic renal cell carcinoma? J Clin Pathol 68(1):12–17
Camp RL, Chung GG, Rimm DL (2002) Automated subcellular localization and quantification of protein expression in tissue microarrays. Nat Med 8(11):1323–1328
Cerami E et al (2012) The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2(5):401–404
Gao J et al (2013) Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 6(269):pl1
Hoadley KA et al (2018) Cell-of-origin patterns dominate the molecular classification of 10,000 tumors from 33 types of cancer. Cell 173(2):291-304.e6
Bi K et al (2021) Tumor and immune reprogramming during immunotherapy in advanced renal cell carcinoma. Cancer Cell 39(5):649-661.e5
Iwata T et al (2021) A new bioinformatics approach identifies overexpression of GRB2 as a poor prognostic biomarker for prostate cancer. Sci Rep 11(1):5696
Blessin NC et al (2019) Patterns of TIGIT expression in lymphatic tissue, inflammation, and cancer. Dis Markers 2019:5160565
Pescia C et al (2023) TIGIT in Lung cancer: potential theranostic implications. Life 13(4):1050. https://doi.org/10.3390/life13041050
Kluger HM et al (2017) PD-L1 Studies across tumor types, its differential expression and predictive value in patients treated with immune checkpoint inhibitors. Clin Cancer Res 23(15):4270–4279
Acknowledgements
The authors would like to acknowledge Lori Charette and Yale Pathology Tissue Services for help in constructing YTMA arrays, and the laboratory of Dr. David Rimm for their generosity in allowing use of their equipment for multispectral image acquisition and analysis. Additionally, we would like to thank the Yale School of Medicine Office of Student Research for their help sponsoring student research for this project.
Funding
The work was funded in part by NIH grants P50 CA121974 (M. Bosenberg and HMK), R01 CA227472 (HMK and K. Herold), R01 CA269349 (HMK), R01 CA269286 (LJ and HMK), R01CA266424 (T. Choueiri and S. Signoretti), K12CA215110 (DAS and HMK), and T32-CA233414-02 (DAS).
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Author contributions are listed below. All authors consent to the publication of this manuscript. D.S., H.K., and O.P. were involved in conceptualization. O.P., D.S., H.K., and D.G.S. assisted with methodology. O.P., D.S., A.A, M.H., D.B., and L.J. conducted the investigation. H.K. acquired the funding. H.K. and D.S. were responsible for project administration and supervision. O.P. D.S., and H.K. contributed to writing-original draft and writing-subsequent drafts.
Corresponding author
Ethics declarations
Conflict of interests
HMK has received consulting fees from Iovance, Merck, Bristol-Myers Squibb, Chemocentryx, Signatero, Gigagen, GI Reviewers, Pliant Therapeutics, Esai, Invox and Wherewolf, all outside of the submitted work. HK has also received research grant funding (to Yale University) from Merck, Bristol-Myers Squibb and Apexigen. MH reports—Advisory Boards: Affini-T, Exelixis, Janssen, CRISPR Therapeutics, Pliant, Regeneron, TScan; Research: Allogene, Alpine, Achilles Therapeutics, Apexigen, Arrowhead, Astellas, AstraZeneca, Bristol Myer Squibb, CRISPR Therapeutics, Eli Lilly, Lyell, Endocyte, Fate Therapeutics, Genentech, Genmab, Iovance, Merck, Novartis, Pfizer, Seattle Genetics, Synthekine, Tmunity, TScan, Xilio; Other: Arvinas, Faeth. DAB reports advisory board fees from Exelixis, AVEO, Eisai, and Elephas, equity in Elephas, Fortress Biotech (subsidiary) and CurIOS Therapeutics, consulting/personal fees from Cancer Expert Now, Adnovate Strategies, MDedge, CancerNetwork, Catenion, OncLive, Cello Health BioConsulting, PWW Consulting, Haymarket Medical Network, Aptitude Health, ASCO Post/Harborside, Targeted Oncology, Merck, Pfizer, MedScape, Accolade 2nd.MD, DLA Piper, AbbVie, Compugen, Link Cell Therapies, Scholar Rock, and research support from Exelixis and AstraZeneca, outside of the submitted work. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Ethical approval
Specimens and clinical information were collected with the approval of a Yale University Institutional Review Board.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Perales, O., Jilaveanu, L., Adeniran, A. et al. TIGIT expression in renal cell carcinoma infiltrating T cells is variable and inversely correlated with PD-1 and LAG3. Cancer Immunol Immunother 73, 192 (2024). https://doi.org/10.1007/s00262-024-03773-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00262-024-03773-8