Abstract
Background
Overall Survival (OS) and Progression-Free Survival (PFS) analyses are crucial metrics for evaluating the efficacy and impact of treatment. This study evaluated the role of clinical biomarkers and dosimetry parameters on survival outcomes of patients undergoing 90Y selective internal radiation therapy (SIRT).
Materials/Methods
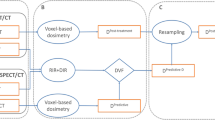
This preliminary and retrospective analysis included 17 patients with hepatocellular carcinoma (HCC) treated with 90Y SIRT. The patients underwent personalized treatment planning and voxel-wise dosimetry. After the procedure, the OS and PFS were evaluated. Three structures were delineated including tumoral liver (TL), normal perfused liver (NPL), and whole normal liver (WNL). 289 dose-volume constraints (DVCs) were extracted from dose-volume histograms of physical and biological effective dose (BED) maps calculated on 99mTc-MAA and 90Y SPECT/CT images. Subsequently, the DVCs and 16 clinical biomarkers were used as features for univariate and multivariate analysis. Cox proportional hazard ratio (HR) was employed for univariate analysis. HR and the concordance index (C-Index) were calculated for each feature. Using eight different strategies, a cross-combination of various models and feature selection (FS) methods was applied for multivariate analysis. The performance of each model was assessed using an averaged C-Index on a three-fold nested cross-validation framework. The Kaplan-Meier (KM) curve was employed for univariate and machine learning (ML) model performance assessment.
Results
The median OS was 11 months [95% CI: 8.5, 13.09], whereas the PFS was seven months [95% CI: 5.6, 10.98]. Univariate analysis demonstrated the presence of Ascites (HR: 9.2[1.8,47]) and the aim of SIRT (segmentectomy, lobectomy, palliative) (HR: 0.066 [0.0057, 0.78]), Aspartate aminotransferase (AST) level (HR:0.1 [0.012–0.86]), and MAA-Dose-V205(%)-TL (HR:8.5[1,72]) as predictors for OS. 90Y-derived parameters were associated with PFS but not with OS. MAA-Dose-V205(%)-WNL, MAA-BED-V400(%)-WNL with (HR:13 [1.5–120]) and 90Y-Dose-mean-TL, 90Y-D50-TL-Gy, 90Y-Dose-V205(%)-TL, 90Y-Dose- D50-TL-Gy, and 90Y-BED-V400(%)-TL (HR:15 [1.8–120]) were highly associated with PFS among dosimetry parameters. The highest C-index observed in multivariate analysis using ML was 0.94 ± 0.13 obtained from Variable Hunting-variable-importance (VH.VIMP) FS and Cox Proportional Hazard model predicting OS, using clinical features. However, the combination of VH. VIMP FS method with a Generalized Linear Model Network model predicting OS using Therapy strategy features outperformed the other models in terms of both C-index and stratification of KM curves (C-Index: 0.93 ± 0.14 and log-rank p-value of 0.023 for KM curve stratification).
Conclusion
This preliminary study confirmed the role played by baseline clinical biomarkers and dosimetry parameters in predicting the treatment outcome, paving the way for the establishment of a dose-effect relationship. In addition, the feasibility of using ML along with these features was demonstrated as a helpful tool in the clinical management of patients, both prior to and following 90Y-SIRT.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Liver cancer is one of the most common cancers worldwide, with a substantially increasing incident rate. Hepatocellular carcinoma (HCC), in particular, stands out as a leading contributor to cancer-related mortality [1, 2]. Selective internal radiation therapy (SIRT) through intrahepatic arterial injection of 90Y microspheres has been used for several decades as a treatment option [3, 4]. The safety and effectiveness of SIRT for primary and metastatic liver cancer with promising post-therapy overall survival (OS) is evident [5, 6]. The treatment has demonstrated more effective results by developing personalized dosimetry [7]. The goal of the treatment is delivering a lethal absorbed dose to the tumors while sparing the healthy liver tissue by tailoring the absorbed dose values to each specific individual [7,8,9,10].
The SIRT procedure is always simulated by similarly injected technetium-99 (99mTc) macro aggregated albumin (MAA) to perform a pre-therapy scan (planar and SPECT/CT). The main objectives are to identify patients with high lung shunt and/or gastrointestinal shunt, who are contraindicated to SIRT, and recently to perform personalized treatment planning and pre-therapy dosimetry [7, 10, 11].
After therapy, the patients undergo follow-up imaging, including multiphasic contrast-enhanced CT or MRI and 18F-FDG PET/CT to evaluate the response to treatment. The response is reflected in changes in total lesion glycolysis measured on follow-up 18F-FDG PET, as well as tumor response as assessed by different versions of Response Evaluation Criteria in Solid Tumors (RECIST) [12,13,14,15,16,17]. The primary clinical objectives of follow-up imaging are the evaluation of tumor local control and early identification of non-responders to the treatment. The aim is to enhance the decision-making process. Nonetheless, the ultimate endpoint is the appraisal of overall survival (OS) [17,18,19].
Many institutions across the world utilize biomarkers, such as baseline tumor stage, liver function, tumor markers and performance status as the cornerstones for prognostication and survival assessment and making decisions for treatment options of an individual accordingly [20]. The OS and progression free survival (PFS) of patients undergoing 90Y SIRT in its different aspects, using these biomarkers has been evaluated in previous studies [21,22,23,24,25,26,27].
Nowadays as dosimetry for 90Y-SIRT patients advances, specifically with developing personalized dosimetry, absorbed dose thresholds are used for treatment planning and treatment verification. These thresholds, known as dose-volume constraints (DVCs) to the tumor and organs at risk (OAR), were largely derived from external beam radiotherapy (EBRT) practices. However, due to significant clinical variations in radiation quality, dose rate, and other factors between EBRT and SIRT, the biological responses and implications of these two treatments is different [28,29,30]. Consequently, and as evidence strongly support the presence of absorbed dose-effect correlation, investigations have delved into establishing an absorbed dose threshold for achieving responses in 90Y-SIRT [7, 31,32,33]. Such correlations indicate that personalized dosimetry-based treatments would improve the outcomes and increase the survival [34].
Recent studies have shown promising results regarding the correlation between tumor mean absorbed dose and OS or PFS [35, 36]. Allimant et al. conducted a comprehensive evaluation of PFS and OS in 45 patients, revealing a significant increase in both parameters with complete tumor targeting (PFS: 2.5 Vs. 7.9 months, OS: 4.5 Vs. 19.2 months) [35]. Similarly, Hermann et al. explored the relationship between tumor absorbed dose and OS in a large cohort from the SARAH trial [36]. Their results indicated that patients who received a mean absorbed dose ≥ 100 Gy with resin microspheres to the tumors exhibited an extended overall survival (14.1 vs. 6.1 months). However, current studies are limited to using statistical approaches for survival analysis considering only the association of tumor absorbed dose and survival without taking into account the corresponding absorbed dose values to normal tissues in this context.
In recent years, there has been increasing interest in applying machine learning (ML) algorithms to survival analysis tasks to assist in determining prognostic indicators [37]. Compared to conventional statistical models that struggle to capture the non-linear relationships between co-variates and output, ML models can learn the multivariate and non-linear correlations present in the training data, thus minimizing the uncertainties, and providing more robust predictions.
We therefore conducted a preliminary, yet comprehensive assessment of the contribution made by clinical biomarkers and established DVCs either for tumors and OARs in predictive modeling of the time-to-event OS and PFS outcomes for patients who underwent 90Y-SIRT procedures. To this end, we developed ML models that utilize the DVCs derived from 99mTc-MAA and 90Y physical and biological effective dose (BED) maps in addition to clinical biomarkers to predict the OS and PFS.
Materials and methods
Patient characteristics
A retrospective study of 17 HCC patients treated with 90Y-glass microspheres (Therasphere™; Boston scientific group, Marlborough, Massachusetts) at Geneva University Hospital (Switzerland), between November 2021 and January 2023 was conducted. Adult patients over 18 years old with at least one tumor > 3 cm, stable liver enzymes, no contraindications to angiography, no concurrent treatment, no previous transplantation, or previous liver resection and an Eastern Cooperative Oncology Group (ECOG) performance status of 0 to 1 were included. All patients underwent personalized dosimetry and treatment planning using 99mTc-MAA SPECT/CT imaging and treatment dose distributions were verified after 90Y treatment by Bremsstrahlung SPECT/CT imaging. Patient characteristics (including clinical biomarkers) are shown in Table 1.
SIRT treatment
Liver vessels were mapped, and extrahepatic shunting was evaluated through angiography. A patient-specific treatment plan was developed based on voxel-wise dosimetry from 99mTc-macroaggregated albumin (99mTc-MAA) SPECT/CT images as a surrogate of 90Y microspheres. Lung shunt fraction (LSF) was calculated based on planar images. Dosimetry and treatment planning was conducted using Simpliciti90YTM treatment planning system (Mirada Medical Ltd, United Kingdom). The median of administered activity of 99mTc-MAA was 156 MBq [range: 133–181]. After an interval of 32.8 ± 20.25 days, a median activity of 2.8 GBq [range: 1.24–6.3 GBq] of 90Y microspheres was administered to the same vessels through catheterization and the residual activity inside the vials was measured.
Image analysis
Multi-phasic contrast-enhanced CT on the SOMATOM Definition Edge (Siemens Healthineers, Erlangen, Germany) or MR images performed on 3T Magnetom Skyra (Siemens Healthineers, Erlangen, Germany, ) were acquired almost a month before simulation (baseline). SPECT/CT images were acquired right after simulation or treatment on a dual-head Symbia-T series camera (Siemens Healthineers, Erlangen, Germany) by imaging 99mTc photons with [128–150] keV energy window using a low-energy, high-resolution collimator, a 128 × 128 matrix, 64 views, over a 180-degree arc and 20–25 s per view. SPECT/CT data were reconstructed using 3D-ordered-subset expectation maximization (3D-OSEM) algorithm with 4 iterations and 8 subsets, while scatter correction was off, attenuation correction was on, and a gaussian postprocessing filter of 5 mm was applied. The images of 90Y-bremsstrahlung photons were acquired under the same scanner and parameters with the following differences: continuous energy window [105–195] keV using high energy collimator, and 64 views with 15–30 s per view.
Dosimetry
The target volumes (tumors) were segmented on diagnostic images and the perfused lobe was delineated on the co-registered attenuation correction CT (ACCT) of SPECT/CT images by an experienced nuclear medicine specialist. Tumor segmentations were transferred on the SPECT/ CT images through registration of diagnostic images and co-registered CT of SPECT/CTs using elastix toolbox through a rigid registration followed by a deformable registration transform. The whole liver was segmented using a previously trained deep learning model with a dice factor of ~ 97% [38] on ACCT of SPECT/CT images, followed by visual check and modification, if necessary.
We calculated the physical as well as the biological effective dose (BED) maps using both 99mTC-MAA simulation and 90Y therapy for each patient as follows:
3D Voxel-wise physical dose maps were calculated according to the Local Deposition Method (LDM, denoted as physical or Dose in this study) by applying an in-house MATLAB code (MATLAB (2022b), Natick, Massachusetts: The MathWorks Inc) validated against replicated analysis with Simplicit90Y™ (Mirada Medical, United Kingdom).
The voxel-wise BED maps were calculated separately for tumor and for healthy structures (normal perfused liver (NPL) and whole normal liver (WNL)) by converting the physical absorbed dose of each voxel to BED using:
where D is the cumulative dose of 90Y radiation, TRep is the sublethal damage repair half-time and Tphys is the radionuclide decay half-life (64.2 h). \({~}^{\alpha }\!\left/ \!{~}_{\beta }\right.\) ratios were set to 10 for both normal and tumoral tissues, whereas TRep (h) was set to 1.5 and 2.5 for Tumoral and Normal structures, respectively. These values were derived from a radiobiological study on glass-microsphere SIRT by Chiesa et al. [39].
The bin size was set to 0.1 Gy to construct dose volume histograms (DVHs). We calculated the Mean absorbed dose, maximum, minimum, DVCs obtained from DVHs including D50, D70, D95, D98, V20, V30, V50, V70, V90, V120, V205, V400 (Dx: minimum dose received by x% of the volume; Vx: the percentage of the volume receiving at least x Gy) as recommended by a body of evidence[33, 40,41,42], for three defined volumes of interest including Tumoral liver (TL), NPL, WNL. The two laters are obtained from subtracting the TL from perfused liver and whole liver structures, respectively. Also, tumor to normal liver ratio (TNR) with respect to the mass of NPL (TNRNPL), and WNL (TNRWNL) was calculated for both simulation and therapy [40]. We also introduced a simplified homogeneity index (HI) borrowed from EBRT; defined as D5/D95 of tumor [43]. Other dosimetry related parameters such as 90Y-injected activity and lung shunt fraction and volume of each structure were included among the features. All dosimetry metrics were calculated once for physical dose maps and once for Biologically effective dose maps.
Endpoints and strategy designing
The endpoints of this study were defined as the time from treatment procedure to death by any cause for OS, and the time from treatment procedure to local disease progression or development of metastasis for PFS.
The biomarkers and dosimetry parameters were considered as predictive features of the OS and PFS endpoints. We designed four distinct strategies for each of the two endpoints, resulting in a total of eight strategies. These strategies are denoted as “Clinical,” “Simulation”, “Therapy,” and “All Features”. The Clinical Strategy is comprised of a total of 16 clinical features. Simulation and therapy strategies include dosimetry features derived from physical and BED dose maps, calculated based on 99mTc-MAA SPET/CT and 90Y SPECT/CT images, respectively. Additionally, both strategies include tumor volume, NPL, and WNL as shape features. All Features Strategy integrates all 305 features utilized across the Clinical, Simulation, and Therapy strategies. The complete name and description of the features are provided in Supplemental-Table 1.
Univariate analysis
To assess the impact of each individual featured on OS and PFS outcomes, a univariate analysis was conducted using Cox proportional hazard models. The Cox models were fitted for each feature to calculate the hazard ratios (HR) and the results were subsequently evaluated using the Wald statistical test. The threshold for statistical significance was set as p-value < 0.05. Additionally, for each feature the concordance index (C-Index) along with corresponding standard errors were calculated. The Kaplan-Meier (KM) curves were constructed for each individual feature. Log-rank test (significance threshold: p-value < 0.05) was used to evaluate the statistical significancy of the observed differences in survival distribution. The median of each continuous feature was considered as the cut-off for group stratification. Noteworthy, the non-usual DVCs such as V30(%)-tumor or D95(Gy)-WNL were not used for univariate analysis.
Feature selection and machine learning modeling
A cross-combination of various supervised ML algorithms and feature selection (FS) methods were analyzed. To identify the most relevant predictors and eliminate redundant features, we employed five different FS methods: univariate C-Index (UCI), Minimal Depth (MD), Mutual Information (MI), Variable hunting (VH), Variable hunting Variable Importance (VH. VIMP).
Using each FS method, No more than seven features were selected based on the recommendation of having a minimum ten observations per feature [44]. The redundant features were removed using Spearman’s correlation test. Pairs of features with a Spearman’s rank correlation coefficient exceeding 0.95 (rho > 0.95) were identified as redundant pairs, and subsequently one from each pair was removed.
The performance of five ML algorithms was evaluated: Cox Proportional Hazard regression (CoxPH), Generalized Linear Model Boosting (GLMB), Generalized Linear Model Network (GLMN), Random Survival Forest (RSF), and Survival Tree (ST). Further details about the FS and ML algorithms utilized in this study can be found in the Supplemental ML section as well as in [45, 46].
We adopted 3-fold cross-validation (CV), whereby there is a combination of inner and outer CV loops. In the inner CV, the dataset was divided into three folds and the model trained on different combinations of training and validation sets. The process was iterated and then the optimal hyperparameters were selected based on the calculated C-indices in the inner CV loop. The outer CV loop was used for estimating the model’s performance; the data were split into three-fold and for each fold, the model was trained on the training dataset using the most effective hyperparameters. This process was iterated for each fold in the outer CV loop, and the C-indices averaged to provide a more robust estimation of the model’s generalization performance. We used this approach to obtain a more reliable performance and to prevent overfitting the hyperparameters to a specific dataset. It should be mentioned that prior to FS and modeling, the features were normalized to their Z-Score based on the training dataset and the same transformation applied to the test dataset of the outer CV loop. Then, FS was performed on the training dataset of the outer CV loop, and the selected features were fed into each model.
In addition, bootstrap aggregation strategy (with 1000 bootstrap samples, with replacement) was implemented to compensate for small data size and prevent overfitting. The Wilcoxon signed-rank statistical test was employed to compare the predictive capabilities of each model. A p-value < 0.05 was considered significant. FS, modeling, and KM curves were calculated using R package (version 4.1). Figure 1 summarizes the adopted methods
Results
Study population characteristics
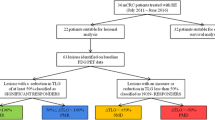
Among all 17 participants in this study, no severe adverse events were recorded. Predominantly, patients exhibited partial responses according to the modified Response Evaluation Criteria in Solid Tumors (mRECIST) criteria. Among the 17 patients included in this study, two patients underwent lobectomy by surgery after 7- and 8-months post-SIRT and were subsequently lost to more follow-up.
The median OS was eleven months with a 95% CI of [8.5, 13 months], ranging from 5 to 20 months. Median PFS was seven months with a 95% CI of [5.6, 10.9 months], ranging from 3 to 20 months.
All patients were followed-up on average 10.8 months (range 6–20 months) after treatment. Two patients failed to follow-up after 7–8 months as they underwent surgical lobectomy and fell out of the follow-up for this treatment. Over the course of 20-months follow up, seven patients passed away, leaving ten cases still alive. Additionally, ten cases developed metastasis or local progression, leaving seven cases free from disease progression. Among ten patients who showed progression after treatment wherein metastasis in the adrenal gland, lymph nodes, subcutaneous and liver metastasis and 6 cases of local progression were observed.
Dosimetry results
The median ± SD of the physical and biologically effective dosimetry parameters for tumors and OARs (NPL and WNL) from both 99mTc-MAA and 90Y are provided in Supplemental-Table 2. The median of LSF was 4.3% [range: 0.8–16.9%]. The median of tumoral mean absorbed doses ± SD were 345.6 ± 179.6, and 662.95 ± 521.65 Gy for 99mTc-MAA physical and BED, respectively. For 90Y, the median of tumoral mean absorbed doses ± SD were 256.23 ± 143.66 and 413.78 ± 583.73 Gy for physical and BED, respectively.
A representative clinical example of physical dose maps and corresponding DVH and BVH (DVH from BED) for 99mTc-MAA and 90Y is demonstrated in Fig. 2.
A 67-year-old male patient diagnosed with HCC who received 2.39 GBq of 90Y as palliative treatment. His OS and PFS were recorded as 6 months. Panel (A) depicts an axial slice of 99mTc-MAA treatment planning dose map along with the corresponding extracted DVH and BVH (DVH form BED). The lesion and normal liver are delineated in red and green, respectively. Panel (B) shows the same information from 90Y treatment verification
Univariate analysis
Figure 3 displays a forest map presenting hazard ratios for statistically significant features (p-value < 0.05) determined through univariate Cox proportional hazards analysis. The results for OS and PFS are reported in the following sections below.
Univariate analysis-OS
Notably, among the clinical features, the presence of “Ascites” revealed a substantial association with OS (HR = 9.2, 95% CI: [1.8, 47]), indicating a significantly higher risk in the Ascites group compared to those without Ascites.
Within the 99mTc-MAA dosimetry features, “MAA-Dose-V205(%)-TL” demonstrated a hazard ratio of 8.5, 95%CI: [1,72]. The comparison groups were stratified based on the median values of the features. Seemingly, patients whose 78% of their tumor was covered by at least 205 Gy demonstrated prolonged OS. Additional details on the univariate Cox proportional hazard models predicting OS can be found in Supplemental-Table 3.
Univariate analysis-PFS
Based on this dataset, no clinical feature was recognized for predicting PFS through univariate analysis. Among 99mTc-MAA dosimetry features, “MAA-Dose-V205(%)-WNL” and “MAA-BED-V400(%)-WNL” were strongly associated with PFS, showing a hazard ratio of 13 (95% CI: [1.5, 120]). Among 90Y-dosimetry features, “90Y-Dose-mean-TL(Gy)”, “90Y-Dose-D50-TL(Gy)”, “90Y-Dose-V205(%)-TL”, “90Y-BED-D50-TL(Gy)”, “90Y-BED-V400(%)-TL” emerged as statistically significant predictors for PFS, with the maximum HR of 15 (95% CI: [1.8, 120]). Patients whose tumors received a mean dose of 260 Gy or higher exhibited an extended PFS compared to those with a tumor dose (TD) less than 260 Gy. Details of the univariate Cox proportional hazard models predicting PFS are summarized in Supplemental-Table 4.
KM curves for all denoted features in Fig. 3 are provided in Supplemental-Figures 1 and 2 with stratification based on the median values of the continuous features. Figure 4 illustrates the KM curves for clinical and mean absorbed dose features predicting OS, as well as the tumoral mean doses that found statistically meaningful for predicting PFS. To optimize the stratification of these features, we also computed the optimal cut points that yielded the most distinct (stratified) KM curves. The corresponding KM curves based on these optimal cut points can be found in Supplemental-Figure 3. The optimal cut points for Aspartate aminotransferase (AST), MAA-Dose-V205-TL (%), 90Y-Dose-mean-TL(Gy) and 90Y-BED-mean-TL(Gy) were 55 (U/L), 75.2%, 244.7 Gy and 413.78 Gy, respectively.
Panel A) The features associated with OS within the univariate analysis. The aim-of-RT means whether patient was treated as 0. Palliative, 1. Lobectomy and 2. segmentectomy. The presence of Ascites, with 0 and 1 legends indicating the non-presenting-ascites and ascites groups, respectively. The non-presenting-ascites group survived longer than ascites group. MAA-Dose-V205-TL (%): The group whose more than 78% of their tumors received at least 205 Gy survived more. Also, the group with Aspartate aminotransferase (AST) < 54 experienced more OS. Panel B) 90Y-tumoral mean dose values from physical and BED calculations associated with PFS; the individuals received mean dose ≥ 260 and mean BED≥ 410 Gy had a longer PFS
Machine learning
The heat map in Fig. 5 displays the mean C-indices resulting from an extensive analysis involving 25 cross-combinations of feature selection and ML models. Accompanying this visualization are the plots showing the results of Wilcoxon statistical tests (p-value) for each strategy. Optimal prognostic models were identified based on both high performance (highest C-indices) and statistical significance. Table 2 summarizes the C-indices, SD, and confidence intervals (CIs) for these selected models.
The KM curves were constructed to visualize the performance of the selected models. The median of the models’ risk scores served as the cut-off for stratification, and the Log-rank test was applied to assess the statistical significance.
The “OS-Therapy-VH.VIMP-GLMN” model exhibited one of the highest C-index values and demonstrated a statistically significant KM curve. Figure 6 shows the KM curves for models that efficiently stratified patients into two groups, regardless of their performance in terms of C-index. Additionally, we provided the KM curves for the selected high C-index models in Supplemental Fig. 4.
The KM curve for models predicting A “OS” and B “PFS” with statistically significant stratification between high and low-risk groups. The calculated median of the risk scores by the models was used as cut-off criteria for stratification. The high-risk individuals had a risk score above or equal to median while the low-risk groups had a risk-score below the median. The p-values were calculated by log-rank statistical test
In Fig. 7, the selected features within the three folds contributing to the predictive power of the OS-Therapy-VH.VIMP-GLMN model are presented. The selected features through each FS method in each strategy are outlined in Supplemental-Tables 6–9 for OS and Supplemental-Tables 10–13 for PFS. Supplemental Figures 5 and 6 depict the feature importance of the selected features, contributing to constructing the efficient (selected) models.
Discussion
Early and accurate prediction of OS and PFS in patients undergoing SIRT procedure is important and can serve as a crucial guide for optimal patient selection, aiding in considering alternative treatments when necessary. The utilization of dosimetry parameters, specifically derived from voxel-level dose maps, contributes to the assessment of treatment planning and verification. These parameters offer insights into the status of tumors and OARs. While the impact of clinical biomarkers and mean absorbed dose in tumor response and OS has been explored through statistical models, there remains a notable gap in understanding the role of other potentially valuable dosimetry parameters in predicting outcomes. Further investigation is imperative to uncover the full spectrum of benefits these features may offer in enhancing predictive models for patient prognosis. Notably, as artificial intelligence plays a pivotal role in medical decision-making, the development of effective predictive models becomes more essential. Establishing robust models for outcome prediction not only facilitates decision-making processes but also enhances accuracy in prognosis. Hence, it’s important to thoroughly explore different dosimetry parameters and include them in detailed predictive models to predict patient outcomes.
We explored the impact of each baseline clinical biomarker, alongside the dosimetry parameters, derived from both physical and biological effective dose maps in treatment planning and verification of SIRT using statistical models. In parallel, 200 models were trained, incorporating various feature selection methods and ML algorithms. These models were designed to predict OS and PFS in four distinct strategies.
This study strategically utilized well-established features extracted from DVH and BVH of the 99mTc-MAA simulation and 90Y treatment verification, instead of additional radiomic or dosiomic features requiring extra time and effort, given the challenges associated with the interpretability of many such features as well as reproducibility over different calculation parameters. Given the lack of spatial information in DVHs, this study does not include the spatial information and the locations where inhomogeneities occur, the information that radiomic and dosiomic features may provide. For instance, we used a homogeneity index defined in EBRT, whose appropriateness for SIRT should be assessed. However, there are some radiomic features that may be more beneficial in this context.
Different scenarios were considered, including the use of only simulation or therapy dosimetry features, only clinical biomarkers, or a comprehensive set of all features combined. This study design allows us to explore and evaluate distinct sets of features tailored to each strategy, leading to understanding of their respective impacts on the study’s endpoints.
To the best of our knowledge, this study is the first study using ML in conjunction with various dosimetry parameters derived from both simulation and therapy sessions. This approach aimed to predict the OS and PFS outcomes in patients undergoing 90Y-SIRT. We extended our analysis to include dosimetry features extracted not only from tumors but also from healthy organs (NPL and WNL). This approach was inspired by a recent study that highlighted the importance of radiomics features from both tumors and organs at risk for predicting OS [46]. To date, a study on SARAH trial dataset indicated that the tumor mean-absorbed-dose (with cutoff of 100 Gy using resin microspheres) from MAA is an independent predictor of prolonged survival [36]. Using glass microspheres, our univariate analysis has unveiled the potential of several features capable of independently predicting OS and PFS (see Fig. 3 and Supplemental Figures 1–2). Our research indicated a mean absorbed dose of ≥ 260 Gy to the tumor, calculated based on 90Y, as a threshold of predicting PFS while in the literature almost the same value was reported for predicting OS [31, 47, 48]. No corresponding value was found as a threshold based on MAA- dosimetry. Nevertheless, it’s crucial to note that the threshold identified in this study should not be regarded as threshold for an optimal clinical outcome.
DVHs are widely used to summarize and quantify dose distribution information, by visually depicting the distribution. However, the granularity of a DVHs depends on the selected dose-volume bin size, and slight changes in bin size can change the shape of the DVH curve. This matter could potentially influence the extracted DVC parameters and the interpretations [49, 50], which should be considered. We used an empirical bin size of 0.1 Gy to consider the minimum detectable changes within a dose distribution. It should be emphasized that we conducted volume-constraint calculations based on both percent and milliliter (ml). The percentage-based approach provides a sense of normalized volume to the total volume. Additionally, we adopted the median of these features as the threshold for stratifying KM curves. For certain features, such as mean absorbed dose values, we adopted the optimal thresholds instead of median. The first (median cutoffs) approach yielded a more balanced distribution of the population between the two groups. Whereas, identifying optimal cutoffs for certain features led to generating an unbalanced distribution.
We evaluated the role of baseline laboratory values, such as albumin, bilirubin, Alpha fetoprotein (AFP), Portal vein thrombosis (PVT), etc. as significant factors for predicting OS [22, 24]. There was no statistically meaningful correlation between AFP or PVT and the endpoints based on our univariate results. However, these features were selected multiple times through feature selection methods demonstrating their predictive importance (Supplemental-Tables 6–13).
Baseline clinical features, including Ascites (log-rank p-value of 0.002) and the aim of Radioembolization, and AST levels were found to be statistically meaningful prognosticators for OS. The aim of Radioembolization including segmentectomy, lobectomy or palliative groups showed a longer OS for segmentectomy group (log-rank p-value = 0.015). An AST level of 54 U/L stratified the KM curves (log-rank p-value = 0.01). The other important baseline clinical prognosticators could be e.g., Barcelona Clinic for Liver Cancer (BCLC), Child Pugh, and performance status (e.g. ECOG). Information regarding BCLC and Child Pugh was missing due to the retrospective nature of the study, and no correlation was observed between ECOG and both endpoints. Additionally, tumor response could serve as an important predictor of survival. However, it was not included as a feature both in univariate and into the models, given the lack of standardized approach for response assessment. Moreover, the primary objective of the study was early prediction of the endpoints using baseline clinical or dosimeric parameters, preferably prior to treatment or based on treatment verification images, excluding follow-up images. Moreover, the impact of tumor response in predicting such outcomes could be found elsewhere [25].
The α/β ratios, which are essential radiosensitivity parameters used for BED calculations in this study, have been sourced from the study by Chiesa et al. [39] where a tumor control probability (TCP) model was developed to extract α values. However, it’s important to note that studies focusing on TCP modeling and biologically effective dosimetry for SIRT are scarce. Many of the studies have utilized values derived from EBRT, which is a fundamentally distinct treatment from SIRT [42]. The EBRT-adopted α and β values are derived from empirical analyses of linear quadratic models, incorporating uncertainties [51]. Therefore, given the inherent differences between EBRT and SIRT, thorough studies on TCP modeling, BED calculations and radiosensitivity parameters are still required.
Our multivariate analysis by ML demonstrated the feasibility of predicting the endpoints using the introduced features. Cross-combination of 25 models and FS methods for 8 strategies resulted in a total of 200 models. The performance of these models was systematically evaluated through the average C-index on a 3-fold CV. Publicly available ML algorithms were selected. They proved to be capable of handling the continuous time-to-event survival data. The feature selection methods were applied to select the most relevant features and to improve the performance of the models. It should be noted that, in the multivariate analysis, we used all the extracted features prior to feature selection, while some of the features might not be evaluated in current (clinical and research-based) dosimetry calculations (e.g. V30 of tumor or D95 of WNL). Nonetheless, we used them all and let the FS algorithms decide on keeping the feature or identifying it as irrelevant or redundant. Noteworthy, for univariate analysis, we did not utilize such features.
While, for each strategy, at least one model outperformed the others in terms of the C-Index (Table 2), we constructed KM curves to validate the overall model performance. Notably, among the KM curves, only one model (OS-Therapy-VH.VIMP-GLMN, C-index = 0.93) that predicted OS using Therapy strategy features effectively stratified the high and low-risk groups (log-rank p-value = 0.023). Although the OS-Clinical-VH.VIMP-GLMN model’s C-index was 0.94, i.e. higher than VH.VIMP-GLMN, this model failed to stratify the groups within KM curves. To explain this phenomenon, C-index assesses how well patients are ranked based on their risk of experiencing an event, whereas the KM curve shows actual survival probabilities over time. While models with high C-index values may effectively rank patients, they may not accurately estimate survival probabilities at specific time points. Therefore, we evaluate models using multiple metrics to ensure performance in different aspects. Limited data may contribute to this discrepancy, as models may excel at calculating the C-index but struggle to predict survival dynamics accurately.
To ensure that ML models’ decision-making procedures are explainable, it’s crucial to clarify the contribution of each individual input feature [52]. Therefore, we provided the importance of the features selected and used by the efficient models (Fig. 7 and Supplemental-Figures 5 and 6).
While our study provided valuable insights, caution should be taken in interpreting the results due to the inherent limitations. The retrospective nature of the study limited our access to only a subset of clinical data, excluding variables, such as Child Pugh and tumor staging, etc. The limited sample size collected from a single center further restricted us in compromising the robustness of the ML models. Another limitation is the lack of external validation. Unfortunately, we did not have access to any clinical database that can serve as external validation dataset to evaluate the robustness of our models on data from other centers with different dose calculation schemes, scanning protocols and population demographics. Moreover, the presence of right censoring in the dataset implies that not all patients had sufficient follow-up time to experience the events of interest. Additionally, the relatively short duration of the follow-up period represents another limitation. Therefore, more investigation on large-scale dataset collected from multiple centers with a long-enough follow-up period is still required.
Although we utilized SPECT images corrected for physical degrading factors, it should be emphasized that the drawbacks of SPECT imaging, such as limited spatial resolution and high noise, effects of physical degrading factors such as attenuation, scatter, and collimator septal penetration etc. may affect the dosimetry results. The images should be corrected for the degrading factors prior to dosimetry calculations. Nevertheless, PET/CT images are more suitable for dosimetry and further studies.
All in all, developing models with large-scale and multiscale dataset, with reproducible results, enables clinicians to tailor individualized treatment strategies and optimize patient care. Identifying patients with good/poor prognosis may affect clinician’s decisions in treatment and follow-up regimen. Patients presenting with good prognosis may be considered for less aggressive treatments or less frequent monitoring, while patients presenting with poor prognosis may benefit from more intensive therapeutic interventions or more frequent follow-ups.
Conclusion
The feasibility of developing ML models to predict survival outcomes using dosimetry parameters obtained from both 99mTc-MAA and 90Y SPECT images, was evaluated. The findings are based on a personalized dosimetry-based dataset that can contribute not only to the refinement of predicting outcomes for SIRT but also has the potential to pave the way for the establishment of dose-effect relationship in SIRT treatment.
Data availability
The data used in this work is not available.
Abbreviations
- TL:
-
Tumoral liver
- NPL:
-
Normal perfused liver
- WNL:
-
Whole normal liver
- OS:
-
Overall survival
- PFS:
-
Progression Free survival
- DVH:
-
Dose volume histogram
- DVC:
-
Dose volume constraints
- ML:
-
Machine learning
- FS:
-
Feature selection
- CV:
-
Cross validation
- SIRT:
-
Selective internal radiation therapy
- SPECT/CT:
-
Single photon emission/ computed tomography
- PET/CT:
-
Positron emission tomography/computed tomography
- MAA:
-
Macro aggregated albumin
- OAR:
-
Organ at risk
- EBRT:
-
External beam radiation therapy
- BED:
-
Biologically effective dose
- HCC:
-
Hepatocellular carcinoma
- NET:
-
Neuro endocrine Tumor
- RE:
-
Radioembolization
- AST:
-
Aspartate aminotransferasie
- ALT:
-
Alanine aminotransferase
- AFP:
-
Alphafeto protein
- LSF:
-
Lung shunt fraction
- 3D-OSEM:
-
3D-ordered-subset expectation maximization
- ECOG:
-
Eastern cooperative oncology group
- ACCT:
-
Attenuation corrected computed tomography
- MAD:
-
Mean absorbed dose
- TNR:
-
Tumor to normal ratio
- UCI:
-
Univariate C-Index
- MD:
-
Minimal Depth
- MI:
-
Mutual Information
- VH:
-
Variable hunting
- VH. VIMP:
-
Variable hunting Variable Importance
- CoxPH:
-
Cox Proportional Hazard regression
- GLMB:
-
Generalized Linear Model Boosting
- GLMN:
-
Generalized Linear Model Network
- RSF:
-
Random Survival Forest
- ST:
-
Survival Tree
- CI:
-
Confidence interval
- C-index:
-
Concordance Index
- SD:
-
Standard deviation
- SARAH:
-
Sorafenib versus Radioembolization in Advanced Hepatocellular Carcinoma
References
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Cancer J Clin. 2021;71(3):209–49.
Bosch FX, Ribes J, Díaz M, Cléries R. Primary liver cancer: worldwide incidence and trends. Gastroenterology. 2004;127(5):S5–16.
Lau W, Leung W, Ho S, Leung N, Chan M, Lin J, et al. Treatment of inoperable hepatocellular carcinoma with intrahepatic arterial yttrium-90 microspheres: a phase I and II study. Br J Cancer. 1994;70(5):994–9.
Dancey JE, Shepherd FA, Paul K, Sniderman KW, Houle S, Gabrys J, et al. Treatment of nonresectable hepatocellular carcinoma with intrahepatic 90Y-microspheres. J Nucl Med. 2000;41(10):1673–81.
Salem R, Lewandowski RJ, Mulcahy MF, Riaz A, Ryu RK, Ibrahim S, et al. Radioembolization for hepatocellular carcinoma using Yttrium-90 microspheres: a comprehensive report of long-term outcomes. Gastroenterology. 2010;138(1):52–64.
Sangro B, Carpanese L, Cianni R, Golfieri R, Gasparini D, Ezziddin S, et al. Survival after yttrium-90 resin microsphere radioembolization of hepatocellular carcinoma across Barcelona clinic liver cancer stages: a European evaluation. Hepatology. 2011;54(3):868–78.
Garin E, Rolland Y, Laffont S, Edeline J. Clinical impact of 99m Tc-MAA SPECT/CT-based dosimetry in the radioembolization of liver malignancies with 90 Y-loaded microspheres. Eur J Nucl Med Mol Imaging. 2016;43:559–75.
Lau W-Y, Kennedy AS, Kim YH, Lai HK, Lee R-C, Leung TW, et al. Patient selection and activity planning guide for selective internal radiotherapy with yttrium-90 resin microspheres. Int J Radiat Oncol Biol Phys. 2012;82(1):401–7.
Sangro B, Iñarrairaegui M, Bilbao JI. Radioembolization for hepatocellular carcinoma. J Hepatol. 2012;56(2):464–73.
Chiesa C, Sjogreen-Gleisner K, Walrand S, Strigari L, Flux G, Gear J, et al. EANM dosimetry committee series on standard operational procedures: a unified methodology for 99m Tc-MAA pre-and 90 Y peri-therapy dosimetry in liver radioembolization with 90 Y microspheres. EJNMMI Phys. 2021;8:1–44.
Levillain H, Bagni O, Deroose CM, Dieudonné A, Gnesin S, Grosser OS, et al. International recommendations for personalised selective internal radiation therapy of primary and metastatic liver diseases with yttrium-90 resin microspheres. Eur J Nucl Med Mol Imaging. 2021;48:1570–84.
Flamen P, Vanderlinden B, Delatte P, Ghanem G, Ameye L, Van Den Eynde M, et al. Multimodality imaging can predict the metabolic response of unresectable colorectal liver metastases to radioembolization therapy with Yttrium-90 labeled resin microspheres. Phys Med Biol. 2008;53(22):6591.
Levillain H, Duran Derijckere I, Marin G, Guiot T, Vouche M, Reynaert N, et al. 90 Y-PET/CT-based dosimetry after selective internal radiation therapy predicts outcome in patients with liver metastases from colorectal cancer. EJNMMI Res. 2018;8:1–9.
van den Hoven AF, Rosenbaum CE, Elias SG, de Jong HW, Koopman M, Verkooijen HM, et al. Insights into the dose–response relationship of radioembolization with resin 90Y-microspheres: a prospective cohort study in patients with colorectal cancer liver metastases. J Nucl Med. 2016;57(7):1014–9.
Fowler KJ, Maughan NM, Laforest R, Saad NE, Sharma A, Olsen J, et al. PET/MRI of hepatic 90Y microsphere deposition determines individual tumor response. Cardiovasc Interv Radiol. 2016;39:855–64.
Strigari L, Sciuto R, Rea S, Carpanese L, Pizzi G, Soriani A, et al. Efficacy and toxicity related to treatment of hepatocellular carcinoma with 90Y-SIR spheres: radiobiologic considerations. J Nucl Med. 2010;51(9):1377–85.
Chapiro J, Duran R, Lin M, Schernthaner R, Lesage D, Wang Z, et al. Early survival prediction after intra-arterial therapies: a 3D quantitative MRI assessment of tumour response after TACE or radioembolization of colorectal cancer metastases to the liver. Eur Radiol. 2015;25:1993–2003.
Gonzalez-Guindalini FD, Botelho MP, Harmath CB, Sandrasegaran K, Miller FH, Salem R, et al. Assessment of liver tumor response to therapy: role of quantitative imaging. Radiographics. 2013;33(6):1781–800.
Laubender RP, Lynghjem J, D’Anastasi M, Heinemann V, Modest DP, Mansmann UR, et al. Evaluating the agreement between tumour volumetry and the estimated volumes of tumour lesions using an algorithm. Eur Radiol. 2014;24:1521–8.
Riaz A, Salem R. Laboratory and imaging prognostic indicators following arterial locoregional therapies for hepatocellular carcinoma survival. J Vasc Interv Radiol. 2019;30(12):1893–4.
Qaseem Y, Salem R. Observing durable responses and a prolonged survival tail in advanced hepatocellular carcinoma with portal vein invasion treated with Y90 radioembolization. Cardiovasc Interv Radiol. 2020;43:1423–4.
Gao R, Gabr A, Mouli S, Riaz A, Kulik L, Lewandowski RJ, et al. Toxicity and survival of hepatocellular carcinoma patients with hepatitis B infection treated with yttrium-90 radioembolization: an updated 15-year study. J Vasc Interv Radiol. 2020;31(3):401–8. e1.
Miller MD, Sze DY, Padia SA, Lewandowski RJ, Salem R, Mpofu P, et al. Response and overall survival for Yttrium-90 radioembolization of hepatic sarcoma: a multicenter retrospective study. J Vasc Interv Radiol. 2018;29(6):867–73.
Gordon AC, Gabr A, Riaz A, Uddin OM, Abouchaleh N, Ali R, et al. Radioembolization super survivors: extended survival in non-operative hepatocellular carcinoma. Cardiovasc Interv Radiol. 2018;41:1557–65.
Ghosn M, Derbel H, Kharrat R, Oubaya N, Mulé S, Chalaye J, et al. Prediction of overall survival in patients with hepatocellular carcinoma treated with Y-90 radioembolization by imaging response criteria. Diagn Interv Imaging. 2021;102(1):35–44.
Chen JX, Rose S, White SB, El-Haddad G, Fidelman N, Yarmohammadi H, et al. Embolotherapy for neuroendocrine tumor liver metastases: prognostic factors for hepatic progression-free survival and overall survival. Cardiovasc Interv Radiol. 2017;40:69–80.
Mähringer-Kunz A, Steinle V, Kloeckner R, Schotten S, Hahn F, Schmidtmann I, et al. The impact of portal vein tumor thrombosis on survival in patients with hepatocellular carcinoma treated with different therapies: a cohort study. PLoS ONE. 2021;16(5):e0249426.
Lee BQ, Abbott EM, Able S, Thompson JM, Hill MA, Kartsonaki C, et al. Radiosensitivity of colorectal cancer to 90Y and the radiobiological implications for radioembolisation therapy. Phys Med Biol. 2019;64(13):135018.
Gholami YH, Willowson KP, Forwood NJ, Harvie R, Hardcastle N, Bromley R, et al. Comparison of radiobiological parameters for 90Y radionuclide therapy (RNT) and external beam radiotherapy (EBRT) in vitro. EJNMMI Phys. 2018;5(1):1–19.
Högberg J, Rizell M, Hultborn R, Svensson J, Henrikson O, Mölne J, et al. Increased absorbed liver dose in Selective Internal Radiation Therapy (SIRT) correlates with increased sphere-cluster frequency and absorbed dose inhomogeneity. EJNMMI Phys. 2015;2:1–17.
Garin E, Tselikas L, Guiu B, Chalaye J, Edeline J, de Baere T, et al. Personalised versus standard dosimetry approach of selective internal radiation therapy in patients with locally advanced hepatocellular carcinoma (DOSISPHERE-01): a randomised, multicentre, open-label phase 2 trial. Lancet Gastroenterol Hepatol. 2021;6(1):17–29.
Garin E, Rolland Y, Edeline J, Icard N, Lenoir L, Laffont S, et al. Personalized dosimetry with intensification using 90Y-loaded glass microsphere radioembolization induces prolonged overall survival in hepatocellular carcinoma patients with portal vein thrombosis. J Nucl Med. 2015;56(3):339–46.
Gabr A, Riaz A, Johnson GE, Kim E, Padia S, Lewandowski RJ, et al. Correlation of Y90-absorbed radiation dose to pathological necrosis in hepatocellular carcinoma: confirmatory multicenter analysis in 45 explants. Eur J Nucl Med Mol Imaging. 2021;48:580–3.
Strigari L, Konijnenberg M, Chiesa C, Bardies M, Du Y, Gleisner KS, et al. The evidence base for the use of internal dosimetry in the clinical practice of molecular radiotherapy. Eur J Nucl Med Mol Imaging. 2014;41:1976–88.
Allimant C, Kafrouni M, Delicque J, Ilonca D, Cassinotto C, Assenat E, et al. Tumor targeting and three-dimensional voxel-based dosimetry to predict tumor response, toxicity, and survival after yttrium-90 resin microsphere radioembolization in hepatocellular carcinoma. J Vasc Interv Radiol. 2018;29(12):1662–70. e4.
Hermann A-L, Dieudonné A, Ronot M, Sanchez M, Pereira H, Chatellier G, et al. Relationship of tumor radiation–absorbed dose to survival and response in hepatocellular carcinoma treated with transarterial radioembolization with 90Y in the SARAH study. Radiology. 2020;296(3):673–84.
Wang P, Li Y, Reddy CK. Machine learning for survival analysis: a survey. ACM Comput Surv (CSUR). 2019;51(6):1–36.
Salimi Y, Shiri I, Mansouri Z, Zaidi H. Deep learning-assistedmultiple organ segmentation from whole-body CT images. Medrxiv. 2023. https://doi.org/10.1101/2023.10.20.23297331:2023.2010.2020.23297331.
Chiesa C, Mira M, Maccauro M, Spreafico C, Romito R, Morosi C, et al. Radioembolization of hepatocarcinoma with 90 Y glass microspheres: development of an individualized treatment planning strategy based on dosimetry and radiobiology. Eur J Nucl Med Mol Imaging. 2015;42:1718–38.
Riveira-Martin M, Akhavanallaf A, Mansouri Z, Bianchetto Wolf N, Salimi Y, Ricoeur A, et al. Predictive value of 99mTc-MAA-based dosimetry in personalized 90Y-SIRT planning for liver malignancies. EJNMMI Res. 2023;13(1):63.
Garin E, Guiu B, Edeline J, Rolland Y, Palard X. Trans-arterial radioembolization dosimetry in 2022. Cardiovasc Interv Radiol. 2022;45(11):1608–21.
Abbott EM, Falzone N, Lee BQ, Kartsonaki C, Winter H, Greenhalgh TA, et al. The impact of radiobiologically informed dose prescription on the clinical benefit of 90Y SIRT in colorectal cancer patients. J Nucl Med. 2020;61(11):1658–64.
Yan L, Xu Y, Chen X, Xie X, Liang B, Dai J. A new homogeneity index definition for evaluation of radiotherapy plans. J Appl Clin Med Phys. 2019;20(11):50–6.
Peduzzi P, Concato J, Feinstein AR, Holford TR. Importance of events per independent variable in proportional hazards regression analysis II. Accuracy and precision of regression estimates. J Clin Epidemiol. 1995;48(12):1503–10.
Mansouri Z, Salimi Y, Amini M, Hajianfar G, Oveisi M, Shiri I, et al. Development and validation of survival prognostic models for head and neck cancer patients using machine learning and dosiomics and CT radiomics features: a multicentric study. Radiat Oncol. 2024;19(1):12.
Salimi Y, Hajianfar G, Mansouri Z, Sanaat A, Amini M, Shiri I, et al. Organomics: a concept reflecting the importance of PET/CT healthy organ radiomics in non-small cell lung cancer prognosis prediction using machine learning. medRxiv. 2024. https://doi.org/10.1101/2024.05.15.24307393.
Alsultan AA, van Roekel C, Barentsz MW, Smits ML, Kunnen B, Koopman M, et al. Dose-response and dose-toxicity relationships for yttrium-90 glass radioembolization in patients with colorectal cancer liver metastases. J Nucl Med. 2021;62(11):1616–23. https://doi.org/10.2967/jnumed.120.255745.
Salem R, Gabr A, Riaz A, Mora R, Ali R, Abecassis M, et al. Institutional decision to adopt Y90 as primary treatment for hepatocellular carcinoma informed by a 1,000-patient 15‐year experience. Hepatology. 2018;68(4):1429–40.
Mansouri Z, Salimi Y, Akhavanallaf A, Shiri I, Teixeira EPA, Hou X, et al. Deep transformer-based personalized dosimetry from SPECT/CT images: a hybrid approach for [177Lu] Lu-DOTATATE radiopharmaceutical therapy. Eur J Nucl Med Mol Imaging. 2024;61(6):1516–29.
Datta NR, Das KJ, Balasubramanium R, Ayyagari S. Spatial information on dose distribution using multisectional dose-volume histograms. Med Dosim. 1996;21(1):19–22.
Lindsey MJ. Analysis of biologically effective dose for retroactive yttrium-90 trans-arterial radioembolization treatment optimization. CMC Senior Theses. 3103. 2023. https://scholarship.claremont.edu/cmc_theses/3103.
Kovalev MS, Utkin LV, Kasimov EM. SurvLIME: a method for explaining machine learning survival models. Knowl Based Syst. 2020;203:106164.
Funding
Open access funding provided by University of Geneva. This work was supported by the Euratom research and training programme 2019–2020 Sinfonia project under grant agreement no. 945196.
Author information
Authors and Affiliations
Contributions
ZM: Conceptualization, Methodology, Software, Validation, Formal analysis, Investigation, Resources, Data curation, Writing – original draft, Writing – review & editing, Visualization. YS, GH: Conceptualization, Methodology, Validation, Formal analysis, Resources, Writing – review & editing, Visualization. BWN, KL, XG, GA, RA, VG, IM: Conceptualization, Methodology, Validation, Investigation, Data curation, Writing – review & editing, Visualization. HZ: Conceptualization, Methodology, Validation, Investigation, Resources, Writing – review & editing, Visualization, Supervision, Project administration, Funding acquisition.
Corresponding author
Ethics declarations
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. This retrospective study was approved by the institutional ethics committee (CCER ID: 2023-00322) and the requirement to obtain informed consent was waived.
Consent to participate
Informed consent was waived in this retrospective study.
Consent to publish
All authors approved the final version of the manuscript and consent to give the Publisher the permission to publish the work.
Competing interests
None of the authors have affiliations that present financial or non-financial competing interests for this work. V. Garibotto is an Associate Editor for the European Journal of Nuclear Medicine and Molecular Imaging.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Mansouri, Z., Salimi, Y., Hajianfar, G. et al. The role of biomarkers and dosimetry parameters in overall and progression free survival prediction for patients treated with personalized 90Y glass microspheres SIRT: a preliminary machine learning study. Eur J Nucl Med Mol Imaging (2024). https://doi.org/10.1007/s00259-024-06805-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00259-024-06805-8