Abstract
Purpose
Neuroinflammation and microglial activation play an important role in amnestic mild cognitive impairment (MCI) and Alzheimer’s disease. In this study, we investigated the spatial distribution of neuroinflammation in MCI subjects, using spectral analysis (SA) to generate parametric maps and quantify 11C–PBR28 PET, and compared these with compartmental and other kinetic models of quantification.
Methods
Thirteen MCI and nine healthy controls were enrolled in this study. Subjects underwent 11C–PBR28 PET scans with arterial cannulation. Spectral analysis with an arterial plasma input function was used to generate 11C–PBR28 parametric maps. These maps were then compared with regional 11C–PBR28 VT (volume of distribution) using a two-tissue compartment model and Logan graphic analysis. Amyloid load was also assessed with 18F–Flutemetamol PET.
Results
With SA, three component peaks were identified in addition to blood volume. The 11C–PBR28 impulse response function (IRF) at 90 min produced the lowest coefficient of variation. Single-subject analysis using this IRF demonstrated microglial activation in five out of seven amyloid-positive MCI subjects. IRF parametric maps of 11C–PBR28 uptake revealed a group-wise significant increase in neuroinflammation in amyloid-positive MCI subjects versus HC in multiple cortical association areas, and particularly in the temporal lobe. Interestingly, compartmental analysis detected group-wise increase in 11C–PBR28 binding in the thalamus of amyloid-positive MCI subjects, while Logan parametric maps did not perform well.
Conclusions
This study demonstrates for the first time that spectral analysis can be used to generate parametric maps of 11C–PBR28 uptake, and is able to detect microglial activation in amyloid-positive MCI subjects. IRF parametric maps of 11C–PBR28 uptake allow voxel-wise single-subject analysis and could be used to evaluate microglial activation in individual subjects.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Amnestic mild cognitive impairment (MCI) is a transitional stage between preclinical Alzheimer’s disease and dementia. Microglial activation plays a significant role in Alzheimer’s disease, along with amyloid and tau deposition [1,2,3]. Recent PET imaging studies have suggested that microglial activation correlates closely with the severity of dementia [1, 4, 5]. However, imaging microglia has been challenging. The 18-kDa translocator protein (TSPO) is a cholesterol-transporter protein expressed in the outer mitochondrial membrane of microglial cells and astrocytes in the brain. TSPO expression in normal brain is very low, but it increases significantly after trauma and inflammation [6,7,8]. The PET tracer 11C–R-PK11195 PET has been used for over 20 years to assess the level of TSPO expression and microglial activation [9,10,11]. However, 11C–R-PK11195 has a poor signal-to-noise ratio [6, 12]. In an attempt to improve the signal-to-noise ratio, several second-generation TSPO radioligands have been developed. The new-generation TSPO tracers are affected by genetic variability of TSPO binding site induced by the rs6971 single-nucleotide polymorphism [13], and recent studies have demonstrated that tracer signal in the high-affinity binders (HAB) is 25–35% higher than in the mixed affinity binders (MAB) [14, 15]. However, our group has previously demonstrated that results gathered from a TSPO subgroup (HAB or MAB) can be translated to the entire AD and MCI population [16].
11C–PBR28 is a second-generation TSPO PET tracer, and can be used to quantify microglial activation in neurodegenerative disease; however, recent studies have demonstrated discordant results in neuroinflammation in AD or MCI subjects using 11C–PBR28 PET imaging [5, 17,18,19]. In fact, the quantification of TSPO is challenging; the tracer binds not only to microglia (and to a lesser extent to astrocytes) in the parenchyma but also to the endothelium and smooth muscle cells [20,21,22]. Since endothelial TSPO is physically in contact with plasma, its apparent affinity for radioligands is higher than parenchymal TSPO.
Spectral analysis (SA) is a powerful kinetic tool for generating parametric maps of ligand volumes of distribution from brain TACs with a plasma input function. As a spectral technique, it makes no assumptions about kinetic compartments, but identifies heterogeneous kinetic components which represent tracer delivery, vascular binding, and later parenchymal tracer-binding components [22, 23].
In this study, for the first time, we performed spectral analysis on 11C–PBR28 PET dynamic images to generate parametric maps of ligand uptake reflected by area under impulse response function (IRF). We compared the SA parametric mapping to the compartmental models and the parametric mapping using Logan graphic analysis. Finally, we evaluated microglial activation between two cohorts (MCI vs HC) using 11C–PBR28 SA parametric mapping, Logan VT and two tissue compartment models, with a view to identify the most appropriate quantification approach for 11C–PBR28 PET in neurodegenerative diseases.
Materials and methods
Demographics
Thirteen MCI patients and nine age-matched healthy controls (all genetically high binders for TSPO ligands — HAB) were recruited from memory clinics and the Join Dementia Research website. This study was approved by the local and regional regulatory ethics committee (London Riverside Research Ethics Committee - National Health Research Services, Health Research Authority, UK), and the approval for administration of radioactivity was given by ARSAC (Administration of Radioactive Substances Advisory Committee).
Image acquisition
MRI scans were acquired for all subjects with a 3 Tesla SIEMENS 32-channel Verio MRI scanner (MPRAGE; time repetition = 2400 ms, time echo = 3.06 ms, flip angle of 9, inversion time = 900 ms, matrix = 256 × 246). The T1 images were used for co-registration of the PET for ROI analysis, while T2-weighted images were used to exclude any significant white matter microvascular disease.
All subjects underwent 11C–PBR28 PET scans with a SIEMENS Biograph TruePoint PET/CT scanner (axial field = 21.8 cm, transaxial planes =111, and spatial resolution = 2.056 mm × 2.056 mm × 2 mm) at Imanova, London. A low-dose CT scan was performed for attenuation correction, followed by injection of a mean activity of 300 MBq 11C–PBR28 intravenously. A continuous 3D dynamic acquisition was performed in list mode for 90 min. The dynamic 11C–PBR28 PET data were reconstructed using scatter correction, attenuation correction and random correction. Then the data were corrected for decay and rebinned as 26 time frames.
Twenty-one subjects (12 MCI and 9 HC) also had 18F–Flutemetamol PET scans using a Siemens Biograph 6 PET/CT scanner. A mean activity of 182 (±2.5) MBq 18F–Flutemetamol was injected intravenously. PET data were acquired 90–120 min after the injection to generate a static 3D 18F–Flutemetamol PET image. The cerebellum was used as the reference region to create the 18F–Flutemetamol RATIO image as previously described [24].
Blood data
All patients had continuous online blood sampling for the first 15 min after the 11C–PBR28 PET scan started, and 12 discrete blood samples were taken at 5, 10, 15, 20, 25, 30, 40, 50, 60, 70, 80, and 90 min, allowing blood and plasma radiotracer activity to be measured. The time course of 11C–PBR28 activity in the plasma was calculated with a linear model fit using plasma-to-blood ratio model in the first 15 min of online whole-blood data. Parent tracer and metabolite levels were measured using HPLC analysis for discrete blood samples. A sigmoid model was applied to describe the parent fraction of 11C–PBR28, which was then used to generate the parent fraction of plasma input function.
Quantification of 11C–PBR28 PET using spectral analysis
Spectral analysis applies a positively constrained general linear model to fit the tissue kinetics with a large matrix of exponential functions with a range of decaying factors convoluted with the plasma input function [25]. The solution to each fit is a vector of linear coefficients αi (peak height), each corresponding to a decay βi (peak position), and is obtained using non-negative least squares (NNLS) [23, 26]. In the solution vector, the non-zero coefficients for the high-frequency components (e.g., large βi) usually reflect the dynamics of the tracer in the blood; the ones with slower βi reflect the kinetics of reversible parenchymal perfusion between plasma or tissue compartments, while the slowest components represent irreversible trapping. The beta-min was used as the decay constant of the 11C (0.00056629) for 11C–PBR28. The tissue 11C–PBR28 ligand-binding response could be measured as portion of impulse response function (IRF), which was reflected by the sum of the intermediate and low frequency components of the spectrum. In this study, we generated IRF(t) parametric maps with different epochs of observation-time: 30 min, 45 min, 60 min, 75 min, and 90 min for 11C–PBR28 using the formula:
where t represents the selected observation time, n denotes number of PET time frames, αi and βi are the peak height and peak position for time frame i. We used a decay constant λ of 0.00056629 for 11C–PBR28. In order to provide a high quality IRF(t) parametric maps, the coefficient of variation (CV) (standard deviation/mean) of 11C–PBR28 binding in major cortices (frontal, temporal, parietal, and occipital lobe) and thalamus were calculated at different observation time; the lower CV indicated a higher precision and lower noise level for that IRF(t) parametric map. (Online Resource 1A).
Quantification of 11C–PBR28 PET using compartmental modelling
Compartmental modelling of TACs was performed to determine the volumes of distribution of the 11C–PBR28 tracer using the parent plasma input function and dynamic PET acquisition. In this study, we fitted kinetic data to two compartmental models for 11C–PBR28 [21, 27]: the two-tissue compartmental model (2TCM4k), and the two-tissue compartmental model with an extra vascular component (2TCM4k-1 K), which accounts for the extra binding compartment for the endothelial cells of blood vessels [21, 28]. We calculated the Akaike information criterion (AIC) to evaluate the model performance [29, 30]. The model with lower AIC was chosen as the preferred model for a predefined ROI. Total volume of distribution (VT), rate constant (k), and binding potential (BPND) were the parameters derived from the kinetic models using MICK software (modelling input function compartmental kinetics) and MATLAB2014a.
Quantification of 11C–PBR28 PET using Logan graphic analysis
Logan analysis is a graphical quantification method which linearises PET uptake data using a plasma or non-specific tissue reference input function where ligands bind reversibly. The gradients of fits represent VT and can be used for parametric mapping at a voxel level, though measurement noise is also linearised and a potential confound. Logan graphic analysis is independent of the number of compartments as long as they are all in equilibrium after a time t. The 11C–PBR28 Logan VT parametric map was generated, where VT reflects the slope of the linear section of the Logan graphic plot [31]. Logan graphical analysis was performed using MICK parametric mapping software developed in MATLAB2014a. CV was measured in major cortices and thalamus to assess the signal variation for the Logan VT.
Region of interest (ROI) analysis in MCI and HC
Regional quantification of mean 11C–PBR28 binding was estimated for compartmental model VT, SA parametric map (IRF) and Logan VT map in MCI and healthy controls. To evaluate the binding, MRI and brain atlas (83-region Hammersmith atlas) were spatially transformed into native PET space using SPM8, and Analyze 11.0 (AnalyzeDirect) was used to sample the regional mean tracer update in predefined ROI regions as described in our previous studies. The ROI regional mean uptake was sampled in the following regions: posterior cingulate, frontal, temporal, parietal, and occipital lobes. ROI analysis was also performed for medial temporal lobe, hippocampus, and thalamus. The regional ROI value was regarded as significantly raised (positive) for that region when it is elevated more than 2SD above the mean of healthy controls. The MCI subjects were considered as 11C–PBR28 positive when at least one cortical region was positive.
Voxel-level analysis between MCI and HC
The parametric maps generated by SA or Logan graphic analysis allowed us to perform voxel-wise comparisons between patients and healthy controls. 1) Parametric maps were co-registered to their corresponding MRIs, 2) normalization and smoothing (6 mm × 6 mm × 6 mm) were applied to the co-registered PET to generate the individual normalized PET in SPM8, and 3) each patient’s normalized parametric map was then compared against a group of controls to generate a single-subject SPM T-map at voxel level, which localised significant pathological increases in each patient. The significant cluster threshold was set at p < 0.05 with an extent threshold of 50 voxels. Family-wise error rate (FWE) was corrected for multiple comparisons. For each subject who had significant clusters of increased 11C–PBR28, those clusters were extracted as individual VOI (volume of interest) maps, and the total volume of VOI was calculated in mm3 in Analyze 11.0.
Brain amyloid status
As the MCI cohorts are heterogeneous, the brain amyloid status for each subject was evaluated from 18F–flutemetamol RATIO images. In this study, an amyloid-positive subject was defined as having uptake in at least one of the large cortices or two separate cortical regions higher than the mean + 2SD of healthy controls.
Statistics
Group mean and standard deviation were calculated in SPSS23 (SPSS, Chicago, IL, USA), for each diagnostic group, and Student’s t-test was used to interrogate significant increases in the tracer uptake. The Pearson correlation coefficient was applied to measure the degree of linear correlation between two groups of variables using SPSS23, and a p-value of < 0.05 was regarded as a significant correlation.
Results
Demographics
The demographic details of 13 MCI and 9 control subjects (aged 54–79 yrs) are detailed in the Online Resource 1B. The MCI cohort demonstrated significantly lower neuropsychometric test scores compared to the age-matched healthy controls.
Brain amyloid status
18F–Flutemetamol scans were performed for 12 MCI and 9 HC subjects. Seven of the 12 MCI subjects had significantly higher amyloid deposition than healthy controls in multiple brain regions, and were classified as amyloid-positive.
Spectral analysis
We identified three peaks along with fractional blood volume of 0.065 (± 0.013) in spectra (Fig. 1a). The high-frequency kinetic spectral component probably corresponded to the vascular TSPO (see Turkheimer et al., 2007 [20]) while the other two probably represented free and bound tracer in parenchyma. The tissue IRF(t) curve is equivalent to the sum of three spectral component peaks (Fig. 1b). Given different sets of observation time for generating IRF parametric map, IRF at 90 min (IRF-90) demonstrated the lowest noise level with global CV mean of 18% (SD = 4%).
a Kinetic spectrum for a healthy control subject, which revealed three different components [at position βi of 7.07e-04 s-1 (blue), 1.58e-03 s-1 (green) and 5.01e-03 s-1 (red) with amplitude αi of 8.35e-04 s-1, 1.03e-03 s-1, and 1.21e-03 s-1] with fractional blood volume (bv) of 0.065 (cyan). b Predicted curves using spectral analysis IRF (impulse response function) for tracer activity (dashed line) which was measured by the sum of three individual components of the spectrum. The blue (KI), green and red curves corresponded to the three component peaks in the spectrum
The 11C–PBR28 IRF parametric map (Fig. 2) generated by spectral analysis provided the opportunity to evaluate each patient for neuroinflammation at a voxel level. In this study, the single-subject analysis detected seven MCI patients (five amyloid-positive and two amyloid-negative) who had significant clusters of elevated microglial activation in frontal gyrus, temporal lobe, parietal gyrus, anterior cingulate gyrus, occipital lobe and in thalamus compared to the healthy control cohort (Fig. 3 and Online Resource 2). Furthermore, VOI analyses were performed on those seven MCIs who had significantly increased 11C–PBR28, which measured volumes of significantly increased 11C–PBR28 binding ranging from 114,982 to 714,801mm3 (Table 1).
Individual IRF-90 and Logan VT parametric maps demonstrated with corresponding MR image. Upper panel displays an MCI patient, middle panel shows a healthy control (HC), and lower panel demonstrates the group-wise average image (Mean) and standard deviation image (SD). The colour bar on the left represents the colour scale used for IRF-90 images, and the colour bar on the right represents the colour scale used for Logan VT images
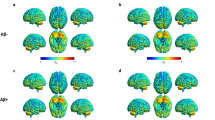
Single-subject VOI analysis of IRF. The colour map represents the significant clusters of increased 11C–PBR28 binding for each MCI patient compared to healthy control cohort. Aβ + and Aβ- represent amyloid-positive MCI and amyloid-negative MCI subjects respectively. The colour bar indicates the significant Z-score which was used for the colour-coded map
In the ROI analysis, parametric mapping of 11C–PBR28 IRF revealed a group-wise significant increase in neuroinflammation in the temporal lobe and multiple other brain regions in the amyloid-positive MCI subjects compared to healthy controls (Table 2). Individually, four amyloid-positive MCIs and two amyloid-negative MCIs revealed ROIs with higher binding than mean + 2SD of healthy controls.
Compartmental modelling
The parent plasma fraction and the plasma:blood ratio of 11C–PBR28 over 90 min are shown in Fig. 4. The 2TCM4k-1 K (AIC = −84) compared with the 2TCM4k model (AIC = −76) provided a marginally better fit of 11C–PBR28 PET uptake data (Online Resource 3). This is consistent with previous studies [21, 32]. VT derived from 2TCM4k-1 K and 2TCM4k correlated well with a Pearson correlation coefficient around 0.7 (p < 0.0001) (Fig. 5).
a The correlation between 11C–PBR28 IRF-90 and Logan VT in frontal lobe and temporal lobe. b The correlation between 11C–PBR28 IRF and 2TCM4k-1 K VT in frontal lobe and temporal lobe. c The correlation between 11C–PBR28 IRF and 2TCM4k VT in frontal lobe and temporal lobe. d The correlation between 2TCM4k-1 K VT and 2TCM4k VT in frontal lobe and temporal lobe
Compartmental VT demonstrated a group-wise increase in thalamus (31%, p < 0.037) and left MTL (32%, p < 0.05) in 11C–PBR28 VT in amyloid-positive MCI subjects compared to the controls. Individually, 4/7 of amyloid-positive MCI subjects showed increased uptake in 11C–PBR28 VT compared to the controls, while one amyloid-negative MCI subject demonstrated an increase in 11C–PBR28 VT. Both compartmental models, 2TCM4k-1 K VT and 2TCM4k VT were positively correlated with the 11C–PBR28 binding in IRF parametric maps (Fig. 5). A Pearson correlation was performed to assess the relationship between 11C–PBR28 IRF-90 and BPND (k3/k4), and a positive correlation was found in frontal lobe (r = 0.55, p = 0.01), temporal lobe (r = 0.7, p < 0.0001), parietal lobe (r = 0.76, p < 0.0001), and occipital lobe ( r = 0.57, p = 0.007). (Online Resource 4).
Logan graphic analysis
Based on linear fit of the whole brain 11C–PBR28 uptake and parent plasma input function, dynamic data from 2000 to 5400 s (six data points) were selected to generate Logan plot (slope = 3.8 ± 0.09 and intercept = −3033 ± 214) to create Logan VT parametric map. Logan VT resulted in a higher CV compared to IRF-90 parametric maps in different cortices (Fig. 2 and Online Resource 1A). With the VOI analysis, Logan maps localised a volume range of 108,237 to 423,979 mm3 containing increased neuroinflammation in individuals (Table 1). At a voxel level, single-subject analysis of 11C–PBR28 Logan parametric VT maps revealed three amyloid-positive MCI subjects and one amyloid-negative MCI subject who had neuroinflammation in anterior cingulate, frontal gyrus, temporal gyrus, parietal lobe, and occipital lobe compared with healthy controls.
A good correlation was found between IRF-90 and Logan parametric maps of 11C–PBR28 (Fig. 5). However, Logan parametric maps failed to localise clusters of significantly raised mean 11C–PBR28 uptake at the group level. Individually, ROI analysis revealed two amyloid-positive and one amyloid-negative MCI subjects with a higher level of 11C–PBR28 binding compared with mean + 2SD of healthy controls.
Discussion
For the first time, we have demonstrated that spectral analysis can be used reliably to quantify 11C–PBR28 PET. 11C–PBR28 PET tracer has 80-fold higher affinity for TSPO compared to previous tracers developed to evaluate microglial activation; however, different methodological approaches used in different studies have produced varying results [5, 14, 15, 33, 34]. As TSPO distribution has a heterogeneous cellular distribution with endothelial, smooth muscle, and parenchymal components all exhibiting different kinetic behaviour, in this study we evaluated the feasibility of generating 11C–PBR28 parametric map using spectral analysis. IRF-90 parametric mapping of 11C–PBR28 correlated well with Logan VT and VT generated by compartment models. Compared to compartmental analysis, the IRF-90 parametric maps provided model free quantification and enable comparison between subjects or between regions at a voxel level. Compared with Logan VT maps, 11C–PBR28 IRF-90 parametric maps showed a lower noise level. IRF parametric mapping of 11C–PBR28 was able to reveal group-wise significant microglial activation in amyloid-positive MCI subjects across multiple brain regions. On an individual basis, IRF-90 parametric maps revealed microglial activation in individuals who had significant amyloid deposition.
As vascular binding can interfere with quantification of parenchymal binding of TSPO tracers [35], spectral analysis [22, 23] has the advantage of separating the high-frequency component which represents tracer binding in the vasculature. SA identifies two tissue components, and another low frequency component which is suggestive of an additional irreversible vascular trapping component to 11C–PBR28 kinetics. Interestingly this is in agreement with the spectral analysis application to 11C–PK11195 [20]. The time-course of changes in 11C–PBR28 IRF is reflected by the sum of low and intermediate frequency components in the spectrum. In this study, we have demonstrated that 90 min is optimal for generating IRF parametric mapping of 11C–PBR28.
It has now been established that TSPO binding is quite heterogeneous within a given area in brain. It is also suggested that TSPO could be expressed in microglia, endothelial cells, and there is a debate about the expression in astrocytes. Some studies have demonstrated that there is irreversible binding to the vascular endothelium, while there is reversible binding to the brain tissue. This gives rise to heterogeneous kinetics of both reversible and irreversible components. Spectral analysis does not make any a-priori assumptions about whether a tracer is reversible or irreversible, and provides optimal parametric map at the pixel level [22, 26]. In the spectrum, each tissue compartment is displayed as a spectral component with an amplitude (α) and a frequency (β). The impulse response function generated by the convolution of these components for 11C–PBR28 PET indicates the response of the brain tissue in response to the injected TSPO tracer. This could be quantified using a complex mathematical model as described below. A spectral analysis allows us to quantify the tissue response throughout the whole cortex without any a-priori assumption. Additionally, it allows us to quantify the tracer uptake in an individual subject. There is extensive evidence in brain imaging and molecular biology studies, which have demonstrated that in-vivo brain amyloid deposition is closely correlated with microglial activation in AD and MCI subjects [36,37,38]. Okello et al. reported that MCI patients with higher amyloid deposition had significantly higher mean levels of cortical microglial activation [39]. In our study, seven MCIs have been classified as amyloid-positive subjects using 18F–flutemetamol PET. Compared to other modelling methods, IRF parametric mapping of 11C–PBR28 has revealed a more extensive profile of neuroinflammation in amyloid-positive MCIs, with a 19–27% group increase across temporal lobe and multiple cortical regions. Compartmental analysis only demonstrated group-wise increased 11C–PBR28 in the amyloid-positive group in the thalamus, while Logan parametric maps failed to localise any significant increase at the group level.
Considering the heterogeneity of neuroinflammation in MCIs, it is crucial for researchers to assess the neuroinflammation on an individual basis rather than as a group. In this study, 11C–PBR28 IRF parametric mapping demonstrated a larger total volume of significantly increased 11C–PBR28, especially for MCI subjects who had amyloid plaques, consistent with the neuropathological findings. Interestingly, one amyloid-positive MCI subject, who revealed a significant cluster of increased microglial activation when IRF and Logan parametric maps were interrogated at a voxel level, was negative with an 11C–PBR28 ROI analysis.
While one of the limitations of the current study is that only MCI subjects have been assessed, we are aiming to recruit established AD patients to further validate the IRF parametric mapping strategy for 11C–PBR28 PET. Despite that, we have previously demonstrated that results gathered from a TSPO subgroup (HAB or MAB) can be translated to an entire AD and MCI population [16]. Another limitation is only HAB cases have been included; our group is recruiting more participants in order to evaluate the performance of 11C–PBR28 IRF in mixed binding affinity groups. SA is highly sensitive and is performed using arterial input analysis, which is the most robust quantitative method of analysis. Small variation could lead to bias in data estimation; however, the arterial blood acquisition and modelling in this study were carefully performed. Nevertheless, this preliminary report has revealed powerful evidence that parametric mapping of 11C–PBR28 using spectral analysis which separates the endothelial component provides a high-quality whole-brain map which could be used to analyze MCI subjects on an individual basis.
Conclusion
In this study, we have demonstrated that spectral analysis can be used to generate 11C–PBR28 parametric maps. We have identified a high-frequency blood volume peak and three other spectral peaks which represent a slow irreversible trapping component and two tissue components. Results demonstrated 11C–PBR28 parametric maps generated by spectral analysis had a signal-to-noise ratio large enough to evaluate the microglial activation at voxel level on an individual basis. In conclusion, we have demonstrated, for the first time, that IRF parametric map generated by spectral analysis can reliably be used in quantifying 11C–PBR28 PET to assess neuroinflammation in prodromal Alzheimer’s disease.
References
Calsolaro V, Edison P. Neuroinflammation in Alzheimer’s disease: current evidence and future directions. Alzheimers Dement. 2016;12(6):719–32.
Heneka MT, Carson MJ, El Khoury J, Landreth GE, Brosseron F, Feinstein DL, et al. Neuroinflammation in Alzheimer’s disease. Lancet Neurol. 2015;14(4):388–405.
Higuchi M, Ji B, Maeda J, Sahara N, Suhara T. In vivo imaging of neuroinflammation in Alzheimer’s disease. Clin Exp Neuroimmunol. 2016;7(2):139–44.
Edison P, Ahmed I, Fan Z, Hinz R, Gelosa G, Ray Chaudhuri K, et al. Microglia, amyloid, and glucose metabolism in Parkinson’s disease with and without dementia. Neuropsychopharmacology. 2013;38(6):938–49.
Kreisl WC, Lyoo CH, Liow JS, Wei M, Snow J, Page E, et al. (11)C-PBR28 binding to translocator protein increases with progression of Alzheimer’s disease. Neurobiol Aging. 2016;44:53–61.
Dickens AM, Vainio S, Marjamaki P, Johansson J, Lehtiniemi P, Rokka J, et al. Detection of microglial activation in an acute model of neuroinflammation using PET and radiotracers 11C-(R)-PK11195 and 18F-GE-180. J Nucl Med. 2014;55(3):466–72.
Su Z, Herholz K, Gerhard A, Roncaroli F, Du Plessis D, Jackson A, et al. [(1)(1)C]-(R)PK11195 tracer kinetics in the brain of glioma patients and a comparison of two referencing approaches. Eur J Nucl Med Mol Imaging. 2013;40(9):1406–19.
Janssen B, Vugts DJ, Funke U, Molenaar GT, Kruijer PS, van Berckel BN, et al. Imaging of neuroinflammation in Alzheimer’s disease, multiple sclerosis and stroke: recent developments in positron emission tomography. Biochim Biophys Acta. 2016;1862(3):425–41.
Ko JH, Koshimori Y, Mizrahi R, Rusjan P, Wilson AA, Lang AE, et al. Voxel-based imaging of translocator protein 18 kDa (TSPO) in high-resolution PET. J Cereb Blood Flow Metab. 2013;33(3):348–50.
Yokokura M, Terada T, Bunai T, Nakaizumi K, Takebayashi K, Iwata Y, et al. Depiction of microglial activation in aging and dementia: positron emission tomography with [11C]DPA713 versus [11C]( R)PK11195. J Cereb Blood Flow Metab. 2017;37(3):877–89.
Fan Z, Brooks DJ, Okello A, Edison P. An early and late peak in microglial activation in Alzheimer’s disease trajectory. Brain. 2017;140(3):792–803.
Alam MM, Lee J, Lee SY. Recent progress in the development of TSPO PET ligands for neuroinflammation imaging in neurological diseases. Nucl Med Mol Imaging. 2017;51(4):283–96.
Owen DR, Yeo AJ, Gunn RN, Song K, Wadsworth G, Lewis A, et al. An 18-kDa translocator protein (TSPO) polymorphism explains differences in binding affinity of the PET radioligand PBR28. J Cereb Blood Flow Metab. 2012;32(1):1–5.
Yoder KK, Nho K, Risacher SL, Kim S, Shen L, Saykin AJ. Influence of TSPO genotype on 11C-PBR28 standardized uptake values. J Nucl Med. 2013;54(8):1320–2.
Park E, Gallezot JD, Delgadillo A, Liu S, Planeta B, Lin SF, et al. (11)C-PBR28 imaging in multiple sclerosis patients and healthy controls: test-retest reproducibility and focal visualization of active white matter areas. Eur J Nucl Med Mol Imaging. 2015;42(7):1081–92.
Fan Z, Harold D, Pasqualetti G, Williams J, Brooks DJ, Edison P. Can studies of neuroinflammation in a TSPO genetic subgroup (HAB or MAB) be applied to the entire AD cohort? J Nucl Med. 2015;56(5):707–13.
Kreisl WC, Lyoo CH, McGwier M, Snow J, Jenko KJ, Kimura N, et al. In vivo radioligand binding to translocator protein correlates with severity of Alzheimer’s disease. Brain. 2013;136(Pt 7):2228–38.
Schuitemaker A, Kropholler MA, Boellaard R, van der Flier WM, Kloet RW, van der Doef TF, et al. Microglial activation in Alzheimer’s disease: an (R)-[(1)(1)C]PK11195 positron emission tomography study. Neurobiol Aging. 2013;34(1):128–36.
Nair A, Veronese M, Xu X, Curtis C, Turkheimer F, Howard R, et al. Test–retest analysis of a non-invasive method of quantifying [(11)C]-PBR28 binding in Alzheimer’s disease. EJNMMI Res. 2016;6(1):72.
Turkheimer FE, Edison P, Pavese N, Roncaroli F, Anderson AN, Hammers A, et al. Reference and target region modeling of [11C]-(R)-PK11195 brain studies. J Nucl Med. 2007;48(1):158–67.
Rizzo G, Veronese M, Tonietto M, Zanotti-Fregonara P, Turkheimer FE, Bertoldo A. Kinetic modeling without accounting for the vascular component impairs the quantification of [C-11]PBR28 brain PET data. J Cereb Blood Flow Metab. 2014;34(6):1060–9.
Veronese M, Rizzo G, Bertoldo A, Turkheimer FE. Spectral analysis of dynamic PET studies: a review of 20 years of method developments and applications. Comput Math Methods Med. 2016;2016:7187541.
Cunningham VJ, Jones T. Spectral analysis of dynamic PET studies. J Cereb Blood Flow Metab. 1993;13(1):15–23.
Hatashita S, Yamasaki H, Suzuki Y, Tanaka K, Wakebe D, Hayakawa H. [18F]Flutemetamol amyloid-beta PET imaging compared with [11C]PIB across the spectrum of Alzheimer’s disease. Eur J Nucl Med Mol Imaging. 2014;41(2):290–300.
Veronese M, Rizzo G, Turkheimer FE, Bertoldo A. SAKE: a new quantification tool for positron emission tomography studies. Comput Methods Prog Biomed. 2013;111(1):199–213.
Turkheimer F, Moresco RM, Lucignani G, Sokoloff L, Fazio F, Schmidt K. The use of spectral-analysis to determine regional cerebral glucose-utilization with positron emission tomography and [F-18] fluorodeoxyglucose — theory, implementation, and optimization procedures. J Cereb Blood Flow Metab. 1994;14(3):406–22.
Zanotti-Fregonara P, Liow JS, Fujita M, Dusch E, Zoghbi SS, Luong E, et al. Image-derived input function for human brain using high resolution PET imaging with [C](R)-rolipram and [C]PBR28. PLoS One. 2011;6(2):e17056.
Guo Q, Owen DR, Rabiner EA, Turkheimer FE, Gunn RN. Identifying improved TSPO PET imaging probes through biomathematics: the impact of multiple TSPO binding sites in vivo. NeuroImage. 2012;60(2):902–10.
Turkheimer FE, Hinz R, Cunningham VJ. On the undecidability among kinetic models: from model selection to model averaging. J Cereb Blood Flow Metab. 2003;23(4):490–8.
Akaike H. Maximum likelihood identification of Gaussian autoregressive moving average models. Biometrika. 1973;60(2):255–65.
Logan J. Graphical analysis of PET data applied to reversible and irreversible tracers. Nucl Med Biol. 2000;27(7):661–70.
Bloomfield PS, Selvaraj S, Veronese M, Rizzo G, Bertoldo A, Owen DR, et al. Microglial activity in people at ultra high risk of psychosis and in schizophrenia: an [C-11]PBR28 PET brain imaging study. Am J Psychiatry. 2016;173(1):44–52.
Lyoo CH, Ikawa M, Liow JS, Zoghbi SS, Morse CL, Pike VW, et al. Cerebellum can serve as a pseudo-reference region in Alzheimer disease to detect neuroinflammation measured with PET Radioligand binding to translocator protein. J Nucl Med. 2015;56(5):701–6.
Guo Q, Owen DR, Rabiner EA, Turkheimer FE, Gunn RN. A graphical method to compare the in vivo binding potential of PET radioligands in the absence of a reference region: application to [(1)(1)C]PBR28 and [(1)(8)F]PBR111 for TSPO imaging. J Cereb Blood Flow Metab. 2014;34(7):1162–8.
Rizzo G, Veronese M, Tonietto M, Bodini B, Stankoff B, Wimberley C, et al. Generalization of endothelial modelling of TSPO PET imaging: considerations on tracer affinities. J Cereb Blood Flow Metab. 2017. https://doi.org/10.1177/0271678X17742004
Fan Z, Aman Y, Ahmed I, Chetelat G, Landeau B, Ray Chaudhuri K, et al. Influence of microglial activation on neuronal function in Alzheimer’s and Parkinson’s disease dementia. Alzheimers Dement. 2015;11(6):608–21.
Gold M, El Khoury J. Beta-amyloid, microglia, and the inflammasome in Alzheimer’s disease. Semin Immunopathol. 2015;37(6):607–11.
Cai ZY, Hussain MD, Yan LJ. Microglia, neuroinflammation, and beta-amyloid protein in Alzheimer’s disease. Int J Neurosci. 2014;124(5):307–21.
Okello A, Edison P, Archer HA, Turkheimer FE, Kennedy J, Bullock R, et al. Microglial activation and amyloid deposition in mild cognitive impairment: a PET study. Neurology. 2009;72(1):56–62.
Funding
This paper presents independent research funded by MRC and supported by the NIHR CRF and BRC at Imperial College Healthcare NHS Trust. The PET scans and MRI scans were funded by the Medical Research Council, and part of the study was funded by Alzheimer’s Research UK.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Dr. Edison was funded by the Medical Research Council and now by Higher Education Funding Council for England (HEFCE). He has also received grants from Alzheimer’s Research UK, Alzheimer’s Drug Discovery Foundation, Alzheimer’s Society, UK, Novo Nordisk, and GE Healthcare. He is also a consultant to Pfizer. Prof. Brooks has received research grants and non-financial support from the Medical Research Council, grants from Alzheimer’s Research Trust, during the conduct of the study; other from GE Healthcare, personal fees from AstraZeneca, personal fees from Cytox, personal fees from Shire, personal fees from Novartis, personal fees from GSK, Holland, personal fees from Navidea, personal fees from UCB, personal fees from Acadia, grants from Michael J Fox Foundation, grants from European Commission, outside the submitted work. Mattia Veronese is supported by the National Institute for Health Research (NIHR) Biomedical Research Centre at South London and Maudsley NHS Foundation Trust and by King’s College London.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Electronic supplementary material
ESM 1
(DOCX 13 kb)
ESM 2
(DOCX 33 kb)
Online Resource 3
AlC values for 2TCM4k and 2TCM4k-1K compartmental model fitting (GIF 43 kb)
Online Resource 4
Correlation between 11C-PBR28 IRF-90 and binding potential (BPND) in frontal lobe, temporal lobe, parietal lobe and occipital lobe. (GIF 30 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Fan, Z., Dani, M., Femminella, G.D. et al. Parametric mapping using spectral analysis for 11C-PBR28 PET reveals neuroinflammation in mild cognitive impairment subjects. Eur J Nucl Med Mol Imaging 45, 1432–1441 (2018). https://doi.org/10.1007/s00259-018-3984-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00259-018-3984-5