Abstract
Blood biochemical indicators play a crucial role in assessing an individual’s overall health status and metabolic function. In this study, we measured five blood biochemical indicators, including total cholesterol (CHOL), low-density lipoprotein cholesterol (LDL-CH), triglycerides (TG), high-density lipoprotein cholesterol (HDL-CH), and blood glucose (BG), as well as 19 growth traits of 206 male chickens. By integrating host whole-genome information and 16S rRNA sequencing of the duodenum, jejunum, ileum, cecum, and feces microbiota, we assessed the contributions of host genetics and gut microbiota to blood biochemical indicators and their interrelationships. Our results demonstrated significant negative phenotypic and genetic correlations (r = − 0.20 ~ − 0.67) between CHOL and LDL-CH with growth traits such as body weight, abdominal fat content, muscle content, and shin circumference. The results of heritability and microbiability indicated that blood biochemical indicators were jointly regulated by host genetics and gut microbiota. Notably, the heritability of HDL-CH was estimated to be 0.24, while the jejunal microbiability for BG and TG reached 0.45 and 0.23. Furthermore, by conducting genome-wide association study (GWAS) with the single-nucleotide polymorphism (SNPs), insertion/deletion (indels), and structural variation (SV), we identified RAP2C, member of the RAS oncogene family (RAP2C), dedicator of cytokinesis 11 (DOCK11), neurotensin (NTS) and BOP1 ribosomal biogenesis factor (BOP1) as regulators of HDL-CH, and glycerophosphodiester phosphodiesterase domain containing 5 (GDPD5), dihydrodiol dehydrogenase (DHDH), and potassium voltage-gated channel interacting protein 1 (KCNIP1) as candidate genes of BG. Moreover, our findings suggest that cecal RF39 and Clostridia_UCG_014 may be linked to the regulation of CHOL, and jejunal Streptococcaceae may be involved in the regulation of TG. Additionally, microbial GWAS results indicated that the presence of gut microbiota was under host genetic regulation. Our findings provide valuable insights into the complex interaction between host genetics and microbiota in shaping the blood biochemical profile of chickens.
Key points
• Multiple candidate genes were identified for the regulation of CHOL, HDL-CH, and BG.
• RF39, Clostridia_UCG_014, and Streptococcaceae were implicated in CHOL and TG modulation.
• The composition of gut microbiota is influenced by host genetics.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Blood biochemical indicators can objectively reflect physiologic or pathological alterations, as well as the metabolic and nutritional status of an animal. Cholesterol (CHOL), triglycerides (TG), and blood glucose (BG) are important indicators affecting the growth performance of chickens through carbohydrate and lipid metabolism. In the bloodstream, cholesterol exists as lipoproteins, including high-density lipoprotein cholesterol (HDL-CH), low-density lipoprotein cholesterol (LDL-CH), and very low-density lipoprotein cholesterol (VLDL-CH). Cholesterol homeostasis is essential for cellular and systemic functions, and elevated cholesterol levels in livestock can negatively impact health and productivity, including egg production and quality, reproduction, and hatchability. Furthermore, high cholesterol levels can increase the risk of fatty liver and arteriosclerosis (Jiang et al. 1990; Luo et al. 2020). Poultry abdominal fat deposition and egg yolk formation depend on plasma triglyceride levels. Elevated triacylglycerol levels are associated with metabolic and physiological disorders such as fatty liver and insulin resistance, which can negatively affect the health and productivity of livestock (Hermier et al. 1984; Arsenault et al. 2010). Blood glucose is a crucial source of energy for animals, and changes in blood glucose levels under pathological or environmental conditions can indicate the overall health status of the organism. Blood glucose is involved in processes such as carbohydrate and lipid metabolism, which impacts the meat quality of livestock and poultry (Choe and Kim 2014).
Numerous studies have demonstrated that blood biochemical indicators are subject to genetic regulation. For instance, Cohen et al. (2005) conducted sequencing of the coding region of PCSK9 and identified two nonsense mutations (Y142X and C679X), which were linked to a 40% reduction of LDL cholesterol in plasma (Cohen et al. 2005). Additionally, many studies investigated the polygenic basis of blood LDL-CH, HDL-CH, and TG levels using genome-wide association studies, and found the cumulative effect of multiple common genes (ANGPTL4, AMAC1L2, and MAFB) contributes to polygenic dyslipidemia (Kathiresan et al. 2009; Teslovich et al. 2010; Li et al. 2020). Voight et al. (2012) performed two Mendelian randomization analyses and revealed that carriers of the LIPG 396Ser allele exhibited higher HDL cholesterol levels (Voight et al. 2012). Moreover, research has indicated that growth traits, such as fat deposition, body weight, feed efficiency, and blood biochemistry traits, are not only influenced by host genetic regulation but also by the host’s “second genome,” i.e., the gut microbiome. The gut microbiota plays a key role in maintaining host health by altering host metabolism and impacting physical functions in both healthy and diseased states (Turnbaugh et al. 2006; Yang et al. 2015). Furthermore, the gut microbiota is capable of regulating numerous metabolic processes in the host, including energy homeostasis, glucose metabolism (Karlsson et al. 2013; Heianza et al. 2019), and lipid metabolism (Hu et al. 2022).
Genomic variations encompass various types of genetic mutations, including single-nucleotide polymorphisms (SNPs), small insertions and deletions (indels), and structural variations (SVs) (Hofmann et al. 2017). SNPs and indels have been widely used in population genetics analysis, molecular-assisted breeding, and identification of disease-related genes in animals (Wen et al. 2021). Furthermore, decades of research have shown that structural variations (SVs), such as large deletions, insertions, duplications, inversions, and translocations, can cause significant perturbations in cis-regulatory regions, resulting in quantitative changes in gene expression and phenotypes (Qin et al. 2021). As a result, SVs play a crucial role in genome diversity and are now widely used in animal breeding (Alonge et al. 2020). Currently, there is limited research on the multi-type genetic variants and multi-segmental microbiota on the regulation of blood biochemical indicators, as well as their interactions in chickens.
In this study, we measured five blood biochemical indicators and 19 growth traits of 206 yellow-feathered dwarf broilers and investigated their correlations. Using the sequencing of host whole-genome and 16S rRNA of their duodenum, jejunum, ileum, cecum, and feces microbiota, we assessed the contributions and co-regulations of host genetics and gut microbiota to blood biochemical indicators. This study will provide new perspectives and insights into understanding the growth, development, and metabolic regulation mechanisms of chickens.
Methods
Animals and sampling
A total of 206 male yellow-feather chickens from Guangdong Wen’s Nanfang Poultry Breeding, Co., Ltd. (Xinxing, China) were used in this study. All chickens are hatched on the same day and fed the same diet. The real-time feed intake and body weight of chickens from 56 to 76 days of age, totaling 21 days, were recorded using a feed intake measurement system, and the body weight gain (BWG), metabolic mid-bodyweight (MMBW). and residual feed intake (RFI) during this period were calculated. RFI was calculated based on the average daily feed intake, average daily gain, and metabolic mid-weight as described by Yan et al. (2019). At 78 days of age, whole blood was collected from each bird via the wing vein using a syringe, and the CHOL, LDL-CH, HDL-CH, TG, and BG contents were detected using a fully automatic biochemical analyzer (Hitachi 7020, Japan) (Yang et al. 2012). The feces were obtained from the cloaca by squeezing the abdomen. All birds were euthanized by cervical dislocation and dissected. The abdominal fat weight (AFW), breast muscle weight (BMW), leg weight (LW), and intramuscular fat (IMF) content were weighed using an electronic balance with an accuracy of 0.1 g, and abdominal fat percentage (AFWP), breast muscle percentage (BMWP), and leg weight percentage (LWP) were calculated. The sebum fat thickness (SFT) and body measurements (including body slant length (BSL), shin circumference (SC), shin length (SL), keel length (KL), and breast width (BrW)) were obtained using a tape (Supplementary Table S1). Immediately after dissection, the content (including chyme and mucosa) was collected from the duodenum, jejunum, ileum, and cecum. And all of the samples were collected in well-labeled 2 mL cryovials and rapidly placed in liquid nitrogen. Blood samples and gut content samples were transferred to a − 80 °C freezer for long-term storage.
DNA extraction and sequencing
Host genome DNA and gut microbial DNA were extracted using the Tiangen DNA Extraction Kit (Tiangen Biotech, Beijing, China) and the QIAamp DNA stool mini kit (QIAGEN, Hilden, Germany), respectively. The host DNA library was paired-end sequenced using the Illumina Hiseq 2500 sequencing system with a length of 150 bp and depth of 10 × to ensure the stability and accuracy of the sequencing results. For microbial and fecal DNA, the region amplified was selected to be the 16S rRNA V4 region, using the specific primers 520F/802R (5’-AYTGGGYDTAAAGNG-3’ and 5’-TACNVGGGTATCTAATCC-3’), and 2 × 300 bp sequencing was performed using the Illumina Miseq sequencing system.
Host genome sequencing data processing
Due to the failure of DNA extraction from the blood genome of one individual, a total of 205 individuals were ultimately used for genomic sequencing data processing and subsequent analysis. The chicken reference genome used in this study is version 6 (Galgal6), which was downloaded from the Ensembl website (http://ftp.ensembl.org/pub/release-106/fasta/gallus_gallus/dna/). The paired-end reads were first quality-controlled using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc) software to remove primers, adapters, and low-quality reads from the library building and sequencing. Reads from the same individual and two libraries were mapped to the chicken reference genome using the default parameters of BWA v.0.7.15 (Clevenger et al. 2015) to obtain a combined SAM file. We used the SAMtools v.0.1.19 tool (Danecek et al. 2021) to sort the mapped SAM files according to the physical location of the reference genome and converted them to binary BAM files. Since PCR amplification led to a portion of reads in the final sequencing results from different clusters of the same read, and these reads were duplicated, the duplicate reads were removed using the Genome Analysis Toolkit (GATK) v.4.2.0.0 (McKenna et al. 2010), followed by file index construction using SAMtools v0.1.19.
Microbial sequencing data processing
The paired-end reads of the microbiota were processed into amplicon sequence variants (ASVs) using the QIIME2 (ver 2020.8) (Bolyen et al. 2019) pipeline. ASV methods infer the biological sequences in the sample before the introduction of amplification and sequencing errors and distinguish sequence variants differing by as little as one nucleotide (Callahan et al. 2017). Raw fastq files were de-multiplexed based on barcode information, and then the low-quality reads were filtered with the following criteria: (1) read lengths < 150 bp; (2) contained ambiguous bases; (3) contained mononucleotide repeats > 8 bp; (4) average quality score < 20. The filtered reads were denoised using the DADA2 (The Divisive Amplicon Denoising Algorithm 2) plug-in in the QIIME2. DADA2 is a software package that models and corrects Illumina-sequenced amplicon errors. DADA2 infers sample sequences exactly, without coarse-graining into OTUs (operational taxonomic units), and resolves differences of as little as one nucleotide (Callahan et al. 2016). The sequences after DADA2 denoising are often referred to as ASVs. To further reduce data noise, this study then performed quality control on ASVs with the following criteria: (1) relative abundance > 10–6; 2) detected in more than one sample. Taxonomy analysis of ASVs based on the Silva 16S rRNA gene database v.138 (Quast et al. 2013) using classify-sklearn in QIIME2.
SNPs and indels calling
Based on BAM files, we performed SNPs and indels calling using GATK v.4.2.0.0. To minimize false positive results, only reads with a mapping quality of more than 20 and base quality of more than 20 were used in this study for subsequent genotyping. The variants calling and genotyping were performed using the HaplotypeCaller and GenotypeGVCFs module in GATK. SelectVariants module was used to extract SNPs or indels, respectively, and only biallelic variants were extracted. Indels smaller than 50 bp were filtered out using the “–max-indel-size 50” argument. The detected SNPs and indels were strictly quality-controlled using the VariantFiltration module. The SNP quality control criteria were (1) QD > 10.0; (2) MQ > 10.0; (3) FS < 60.0; (4) MQRankSum > − 12.5; and (5) ReadPosRankSum > − 8.0; and the quality control standard for indels are (1) QD > 2.0; (2) FS < 200.0; (3) QUAL > 30.0; and (4) ReadPosRankSum > − 20.0. The SNPs and indel dataset were quality controlled using PLINK v.1.9 (Purcell et al. 2007) with quality control criteria of (1) sample detection rate > 95%, (2) SNP/indel detection rate > 95%, and (3) minimum allele frequency > 1%. Then, the dataset that met the criteria was genotype-imputed using Beagle v.4.0 (Browning and Browning 2007). SNPs and indels are annotated with ANNOVAR (Wang et al. 2010) and SnpEff (Yen et al. 2017) respectively. Since there is more than one annotation reported in each variant when using SnpEff, when multiple effects are reported, we took the first-ranked annotation for statistics.
Identification of SVs
SVs were identified through Manta v.1.6.0 (Chen et al. 2016) and DELLY v.0.8.7 (Rausch et al. 2012). The two software called SVs by performing mapped paired-end read and split read analyses and were run with default parameters to detect deletions (DEL), inversions (INV), duplications (DUP), and translocations (TRA). SV call sets from Manta and DELLY were then merged with SURVIVOR v.1.0.7 (Jeffares et al. 2017). This will merge all the vcf files specified in sample_files together using a maximum allowed distance of 1 kb, as measured pairwise between breakpoints, and only to report calls supported by 2 callers and they have to agree on the same type and the same stand of the SVs (Jeffares et al. 2017). The quality control of SVs was using the parameter –maf 0.01 –geno0.05 –mind 0.05 and the annotation used ANNOVAR and SnpEff.
Exploring the association between blood biochemical indicators and growth traits
First, the normality of 5 blood biochemical traits and 19 growth traits were assessed using the Shapiro–Wilk test in the R program v.4.2.2 (Wan et al. 2022), and all phenotypic data was transformed using a Box-Cox normal transformation for subsequent analysis (Supplementary Table S2) (Biscay Lirio et al. 1989; Hernandez-Segura et al. 2019). In order to understand the potential association between blood biochemical indicators and growth traits, we conducted Pearson’s correlation analysis using the psych package in R to calculate the correlation coefficients between the blood biochemical traits and growth traits. P-value < 0.05 after BH correction is a significant threshold. We then performed bivariate REML analysis in GCTA v.1.94 (Yang et al. 2011) to estimate the genetic correlation between the blood biochemical traits and growth traits using independent SNP markers. The genetic correlation is defined as \({r}_{g}= \frac{{\sigma }_{g1g2}}{{\sigma }_{g1}{\sigma }_{g2}}\), where the subscripts “1” and “2” represent the two traits; \({\sigma }_{g1g2}\) is the genetic covariance; \({\sigma }_{g1}\) and \({\sigma }_{g2}\) are the genetic variance of traits 1 and 2, respectively. We used the likelihood-ratio test statistic to test the hypotheses that the genetic correlation coefficient is zero (no genetic correlation) and obtain accompanying P values (Deary et al. 2012).
Assessment of the impact of host genetics on blood biochemical indicators
To estimate the proportion of phenotypic variance that is accounted for by genome-wide SNPs, we utilized the GREML algorithm within the GCTA to estimate the heritability of blood biochemical indicators (Yang et al. 2011; Wen et al. 2019). Subsequently, to investigate the phenotype-related host genetic variation, genome-wide association analysis was performed using univariate linear mixed models (ULMM) in GEMMA v.0.98.5 software (Zhou and Stephens 2012). GEMMA can fit a univariate linear mixed model in the following form:
where y is an n-vector of quantitative traits (or binary disease labels) for n individuals; W = (w1, · · ·, wc) is an n × c matrix of covariates (fixed effects) including a column of 1 s; α is a c-vector of the corresponding coefficients including the intercept; x is an n-vector of marker genotypes; β is the effect size of the marker and is an estimate of the marker additive effect; u is an n-vector of random effects; \({\varvec{\upepsilon}}\) is an n-vector of errors; τ−1 is the variance of the residual errors; λ is the ratio between the two variance components; K is a known n × n relatedness matrix and In is an n × n identity matrix. MVNn denotes the n-dimensional multivariate normal distribution. The significance p-value level between SNPs, indels, or SVs and phenotypes was calculated from a derived score test.
To control the effect of population structure on GWAS analysis, we first constructed relatedness matrices based on different variant types. Specifically, if variants with lower minor allele frequency tend to have larger effects, then the standardized genotype matrix is preferred. Therefore, the relatedness matrices were first estimated from the standardized genotype information of SNPs, indels, and SVs respectively. The relatedness matrices were calculated as follows:
We denote X as the n × p matrix of genotypes, xi as its ith column representing genotypes of the ith variant, \({\overline{x} }_{i}\) as the sample mean and \({v}_{xi}\) as the sample variance of ith variant, and 1n as a n × 1 vector of 1’s. Then, the correlation was calculated for the relatedness matrices composed of different variant types. Since the relatedness matrices constructed based on SNPs and indels have high similarity, and the relatedness matrix constructed based on SVs has low similarity with the other two types with the number of detected SVs is small, it is difficult for SVs to capture complete genetic variation information. Therefore, subsequent GWAS with SNPs and indels were conducted with their own relatedness matrix for correction, while GWAS of SVs were corrected using the relatedness matrix constructed based on SNPs. The effect of population stratification was corrected by adjusting the first five principal components (PCs) as derived from the whole-genome SNPs and indels (Price et al. 2010). To avoid potential false positives in multiple comparisons, the whole-genome significance threshold was adjusted via the Bonferroni adjustment (Sedgwick 2014). For SNPs, we first calculated their valid independent count using simpleM (Wen et al. 2021), which was determined to be 1,535,191. Therefore, we set the significance thresholds as − log10(0.05/1,535,191) = 7.49. For indels and SVs, we set the thresholds as − log10(0.05/1,332,016) = 7.43 and − log10(0.05/13,303) = 5.42, respectively. In addition, the Manhattan plots and quantile–quantile (Q-Q) plots of the MLM for individual traits were implemented in R (Gacesa et al. 2022).
Investigating the correlation between gut microbiota and blood biochemical indicators
To determine the effect of gut microbiota from different intestinal segments on phenotype, we estimated the microbiability of five blood biochemical indicators based on microbiota from different segments of the intestine. The proportion of the total variance explained by the gut microbiota is called microbiability (Wen et al. 2021) and is defined as m2 = σ2m/σ2p, where σ2m is the microbial variance. We first constructed a microbial relationship matrix (MRM) based on the Z-score standardized matrix of ASV relative abundance, and then used the REML algorithm in GCTA to calculate the microbiability (Wen et al. 2019) based on the MRM, The formula for calculating the MRM matrix is as follows:
\({\mathbf{M}}_{s}\) is the MRM of the gut segment s, \({\mathbf{X}}_{s}\) is the normalized relative abundance matrix of the taxa, \({\mathbf{X}}_{s}^{T}\mathrm{ is the transpose of}{\mathbf{X}}_{s}\), and Ns is the number of taxa used to estimate the MRM in gut segments (Wen et al. 2019, 2021).
To investigate the specific families associated with blood biochemical indicators, we performed a correlation analysis between families with detection rates > 10% and blood biochemical indicators. Families with detection rates > 50% were considered as continuous phenotypic traits, and the relative abundances were subjected to inverse normal transformation, where all zero counts were turned into missing values (represented by NAs) (Hughes 2020; Grieneisen et al. 2021). Spearman’s correlation analysis was performed between the transformed relative abundances and blood biochemical indicators. A P-value < 0.05 after false discovery rate (FDR) correction was considered to be significantly correlated. Families with detection rates between 10 and 50% were considered as presence/absence (P/A) phenotypes, with presence (relative abundance > 0) coded as 1 and absence (relative abundance = 0) coded as 0. Then, polyserial correlation analysis was performed for these families. A P-value < 0.05 indicates a significant association between the presence/absence of a family and a blood biochemical indicator. Subsequently, a two-tailed analysis was performed on the families with a correlation coefficient greater than 0.2 among the families with a detection rate > 50%. We sequentially selected the top and bottom 20% individuals based on blood biochemical indicators or microbial abundance, and performed Wilcoxon’s rank-sum test on microbial abundance or blood biochemical indicators (Mahajan et al. 2018). If there were significant differences (P < 0.05) in blood biochemical indicators between high and low microbial abundance groups, as well as significant differences (P < 0.05) in microbial abundance between high and low blood biochemical indicator groups, then an association was considered to exist between the blood biochemical indicator and the microbiota.
Exploring the role of host genetics in regulating gut microbiota
To investigate the impact of host genetics on the gut microbiota, we performed a microbial genome-wide association study (mGWAS) analysis using microbial relative abundance as a phenotype with the GEMMA software. Our mGWAS analysis was based on SNPs, indels, and SV information. We classified the families into continuous and P/A types based on their detection rate. Families with a detection rate greater than 50% were categorized as continuous types, and their relative abundances were transformed using inverse normal transformation before analysis. Families with a detection rate between 10 and 50% were classified as P/A types, with 1 indicating presence (relative abundance > 0) and 0 indicating absence (relative abundance = 0). The P values were adjusted using the Bonferroni method. A significant P-value for continuous traits indicated that the relative abundance of the family was genetically regulated, whereas for P/A traits, a significant P-value indicated that the presence or absence of the family was genetically regulated. Subsequently, in order to further investigate the interplay among host genetics, gut microbiota, and blood biochemical traits, we selected bacterial taxa with significant loci in both SNPs, indels, and SV mGWAS analyses. We then searched for shared loci or genes between mGWAS and GWAS. Additionally, we focused on the significant loci or genes from SNPs, indels, and SV mGWAS within these bacterial taxa. Through gene functional annotation and in combination with the correlation analysis between these bacterial taxa and blood biochemical indicators, we identified genes and bacterial taxa that co-regulate blood biochemical indicators.
Results
Association between blood biochemical indicators and chicken growth traits
In this experiment, 24 traits were measured in chickens, including 5 blood biochemical traits, i.e., total cholesterol (CHOL), low-density lipoprotein cholesterol (LDL-CH), triglycerides (TG), high-density lipoprotein cholesterol (HDL-CH), and blood glucose (BG) and 19 growth traits (Supplementary Table S1). The descriptive statistics were performed for the five blood biochemical traits, and it was observed that their coefficients of variation were between 17.27 and 31.87%, indicating a large variability in this population (Table 1). Pearson’s correlation analysis indicated that CHOL, LDL-CH, and TG were significantly negatively correlated with growth traits such as body weight (BW), abdominal fat weight (AFW), abdominal fat percentage (AFP), metabolic mid-bodyweight (MMBW), body weight gain (BWG), breast muscle weight (BMW), leg weight (LW), shin circumference (SC), and breast muscle weight percentage (BMWP) (Table 2). The correlation among blood biochemical traits was all positive, with the highest correlation coefficient between CHOL and HDL-CH (r = 0.92, Padj < 0.001), followed by that between CHOL and LDL-CH (r = 0.7, Padj < 0.001), suggesting a certain degree of synergistic effect of blood biochemical indicators (Fig. 1, Supplementary Table S3). To examine the extent to which the correlation in these traits can be attributed to genetic factors, we estimated the genetic correlations between blood biochemical indicators and growth traits. The results showed that the genetic correlation coefficients among blood biochemical indicators and growth-related traits were generally consistent with the phenotypic correlations. Specifically, CHOL and LDL-CH were significantly and negatively correlated with BW, AFW, AFP, MMBW, BMW, and SC (rG = − 0.40 ~ − 0.67, P < 0.05) (Supplementary Table S3), suggesting the indicative role of CHOL and LDL-CH on growth traits in chicken.
The correlation between 24 traits. The upper triangle is Pearson’s phenotypic correlation analysis, the lower triangle is genetic correlation analysis. The symbols *, **, and *** represent P values < 0.05, 0.01, and 0.001, respectively. Red indicates negative correlations; blue indicates positive correlations
Genetic regulation of blood biochemical indicators in chicken
After a series of quality control, we finally obtained 3,803,923 SNPs and 1,332,016 indels (2 ~ 50 bp). In addition, a total of 13,303 SVs (> 50 bp) were identified, including 11,583 deletions, 1150 duplications, 552 translocations, 17 conversions, and 6 insertions. First, the effects of these genetic variants were evaluated using SnpEff. Only a small fraction of variants (6.67% of SVs) had significant effects ranging from low to high, while 93.33% of SVs were classified as modifiers (no effects). For SNPs and indels, over 98% were identified as modifiers, indicating that the effects of the majority of variants were negligible (Fig. 2A, Supplementary Table S4). Subsequently, using ANNOVAR to classify the effective variants by type and region, we found that intronic and intergenic variations were much higher than other types, totally accounting for more than 75%. While variations in exons accounted for less than 10%, with 0.30% in indels, 1.67% in SNPs, and 9.01% in SVs (Fig. 2B, Supplementary Table S5). This may be due to the fact that the variations in large segments cover a more extensive region and are more prone to cause exon changes. The integration of variants is shown in Table 3.
To explore the effects of host genetics on phenotypes, we first estimated the heritability of these five blood biochemical traits based on whole-genome SNP variants. The results showed CHOL, LDL-CH, HDL-CH, and BG were heritable, with heritability of 0.23, 0.19, 0.24, and 0.15. However, the heritability of TG is negligible (0.03), indicating that plasma TG levels were highly susceptible to the internal metabolic state of the body (Supplementary Table S6). Then, we performed GWAS analysis for 5 blood indicators. To correct the effect of population structure, we firstly constructed relatedness matrices based on SNPs, indels, and SVs respectively. The results showed that the relatedness matrices constructed by SNPs and indels have a high similarity (within-individual: r = 0.86, between-individuals: r ≥ 0.9). However, their correlation with the matrix established by SVs was relatively weak (within-individual: r < 0.1, between-individuals: r = 0.61 ~ 0.82, Fig. 3). This can be attributed to inadequate sequencing depth and limited detection of SV variations, making SVs unsuitable for constructing the relatedness matrix. As such, subsequent GWAS for SNPs and indels were performed with their own relatedness matrix for correction, while GWAS for SVs were corrected using the relatedness matrix constructed based on SNPs.
Correlations of relatedness matrices based on SNPs, indels, and SVs. The upper triangle represents the correlation of relatedness coefficients within the same individual. The lower triangle represents the correlation of relatedness coefficients between different individuals, with green indicating the correlation of relatedness coefficients between all different individuals, and yellow indicating the correlation between individuals with relatedness coefficients greater than 0.1. The diagonal shows the density plot of relatedness coefficients. r denotes the correlation coefficient of relatedness matrices, with ** indicating a P-value < 0.01
The GWAS analysis was performed on 5 blood biochemical indicators with the genotyping data of SNPs, indels, and SVs, and the results revealed significant loci for their association with CHOL, HDL-CH, and BG. The GWAS analysis for HDL-CH identified 16 SNPs, 5 indels, and 3 SVs that were statistically significant (Fig. 4A). These 16 SNPs and 5 indels were annotated to 12 and 5 genes respectively, with two shared genes, DOCK11 and RAP2C, further suggesting their regulatory roles for HDL-CH. Additionally, two deletions of 94 bp and 578 bp were identified by the SVs-GWAS, located upstream of the NTS gene on chromosome 1 and within the intron of the BOP1 gene on chromosome 2, respectively, both of which were associated with cholesterol regulation (Supplementary Tables S7–S10). For CHOL, a suggestive significant locus rs734932526 on chromosome 4 was detected in SNPs-GWAS and a deletion of 263 bp on chromosome 33 was detected in SVs-GWAS. Gene annotation results indicated that both loci reside on novel genes (ENSGALG00000053344 and ENSGALG00000047565) (Supplementary Fig. S1A, Supplementary Tables S7 and S9); for BG, a total of 21 significant loci (10 SNPs, 6 indels, and 5 SVs) were detected. Among the genes annotated by these loci, GDPD5, DHDH, and KCNIP1 genes were considered as the candidate genes for their involvement in lipid and glucose metabolism (Supplementary Fig. S1B, Supplementary Tables S7, S9, and S10). The above results indicate that blood biochemical indicator levels are jointly regulated by SNPs, indels, and SVs, among which CHOL is regulated by DOCK11, RAP2C, NTS, and BOP1, and BG is associated with GDPD5, DHDH, and KCNIP1 genes.
GWAS analysis of HDL-CH. A The Circular-Manhattan plot of HDL-CH. From the outer circle to the inner circle are the SNPs, indels, and SVs-GWAS. The horizontal black solid and grey dashed lines indicate genome-wide significance and suggestive significance thresholds (for SNPs, significant and suggestive significant thresholds were 3.26 and 6.51; for indel were 3.76 and 7.51; for SVs, significant threshold was 5.42). B The QQ plot of CHOL, HDL-CH, and BG
Regulation of blood biochemical indicators by gut microbiota
The final number of samples used for microbial 16S rRNA sequencing was 1026 due to insufficient fecal sample collection from four individuals (cecum, duodenum, ileum, jejunum: 206, feces: 202). A total of 75,029 features were identified by ASV analysis of sequencing data, and a total of 840 species, 691genera, 365 families, 225 orders, 102 classes, and 41 phyla were obtained (Supplementary Table S11).
To assess the effects of gut microbiota from different intestinal segments on the blood biochemical indicators, the microbiabilities were calculated (Fig. 5, Supplementary Table S12). The microbial regulation patterns of CHOL and HDL-CH were found to be similar, with the duodenal microbiota having almost no regulatory effect (m2 = 0), while microbiota in other intestinal segments exhibited varying degrees of regulatory effects (m2 = 0.12 ~ 0.24), with the cecum playing the most significant regulatory role (m2 = 0.2, 0.24). The regulation of BG and TG is mainly controlled by the jejunal microbiota, with m2 reaching 0.45 and 0.23, respectively. The regulatory effects of each intestinal segment on LDL-CH were weak, with only the cecum and feces having slightly higher effects (m2 = 0.13, 0.15). In summary, CHOL, LDL-CH, and HDL-CH levels are primarily regulated by the cecum, while TG and BG levels are mainly regulated by the jejunal microbiota.
To further identify the specific microbes that associated with blood biochemical indicators, the families with detection rates > 50% were used for Spearman’s correlation analysis. Only the microbes with at least one blood biochemical indicator having a |r|> 0.2 and P < 0.05 were retained, resulting in a total of 33 microbes (7 in duodenum, 3 in jejunum, 12 in ileum, 7 in cecum, and 4 in feces, Supplementary Table S13). As a result, weak to moderate correlations (r = − 0.23 ~ 0.25) between gut microbes and five blood biochemical indicators were observed (Fig. 6A). To validate their correlation, we performed a two-tailed analysis for gut microbes with these 33 microbes. We compared the differences in family abundances between individuals with blood biochemical indicators in the top and bottom 20% (N = 40), as well as the differences in blood biochemical indicator levels between individuals with family abundances in the top and bottom 20% (N = 40) in sequence. Finally, the associations of two families RF39 and Clostridia_UCG.014 in the cecum with CHOL, and Streptococcaceae in the jejunum with TG were confirmed. In specific, the individuals with the lowest 20% of relative abundances of RF39 and Clostridia_UCG.014 in the cecum had significantly higher CHOL levels (P = 0.01, 0.0039) than the top 20% individuals. Meanwhile, those in the lowest 20% of CHOL levels had significantly higher relative abundances of RF39 and Clostridia_UCG.014 (P = 0.008, 0.0082) (Fig. 6B, C). In addition, the TG level was significantly lower in the 20% of chickens with the lowest relative abundance of jejunal Streptococcaceae than in the 20% with higher relative abundance (P = 0.00068). Correspondingly, chickens with low relative abundance of jejunal Streptococcaceae had a lower TG level (P = 0.00013) (Fig. 6D). Therefore, these results suggested that RF39 and Clostridia_UCG.014 in the cecum were associated with CHOL, and jejunal Streptococcaceae was associated with TG. We further analyzed the detection rate and relative abundance of RF39, Clostridia_UCG.014, and Streptococcaceae in different gut segments. The results showed that RF39 and Clostridia_UCG.014 were mainly detected in the cecum, with detection rates of 0.96 and 0.84, and Streptococcaceae was mainly colonized in the duodenum with a detection rate of 0.8. The relative abundances of RF39 and Clostridia_UCG.014 in all intestinal segments were with an average abundance of 0.37% and 1.33%, respectively, while the relative abundance of Streptococcaceae can reach up to 8.47% in the jejunum (Fig. 6E–G).
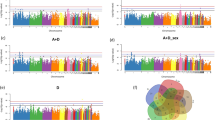
The relationship between families with a detection rate greater than 50% and blood biochemical indicators. A Spearman’s correlation analysis of blood biochemical indicators with the detection rate of > 50% of families. Families with |r|> 0.2 are shown. The upper triangle is the correlation coefficient, and the lower triangle is the P-value (* means P < 0.1, ** means P < 0.05, *** means P < 0.01). B Differences in the CHOL (RF39) between the two groups with the highest and lowest RF39 abundance (CHOL). C Differences in the CHOL (Clostridia_UCG.014) between the two groups with the highest and lowest Clostridia_UCG.014 abundance (CHOL). D Differences in the TG (Streptococcaceae) between the two groups with the highest and lowest Streptococcaceae (TG). E–G Relative abundance and detection rate of RF39, Clostridia_UCG.014, and Streptococcaceae in different intestinal segments. The purple represents the relative abundance, and the blue line represents the detection rate
As to the families with detection rates within 10 ~ 50%, polyserial correlation analysis was performed and retained only the microbes that had a significant correlation coefficient with at least one blood biochemical indicator greater than 0.2 (47 microbes, Supplementary Table S14). The correlation between P/A microbiota and phenotypes was also found to be in the range of low to moderate correlation (− 0.34 to 0.32). The polyserial correlation analysis indicated a generally positive correlation between the presence of families in the ileum with blood biochemical indicators, with the largest correlation coefficient of 0.32 observed between Butyricicoccaceae and BG. Meanwhile, negative correlations were observed in most of the microbes in the duodenum, jejunum, and cecum, with Enterococcaceae in the cecum exhibiting a correlation coefficient of − 0.34 with HDL-CH (Fig. 7A).
Polyserial correlation and mGWAS of P/A families. A Polyserial correlation between blood biochemical indicators and P/A families. The number in the color represents the correlation coefficient and families with |r|> 0.2 are shown. * indicates that significant loci were detected in these families in both SNPs, indels, and SV mGWAS. B The number of families with significant loci detected by SNPs, indels, and SV mGWAS. C The distribution of P/A traits with significant loci in different gut segments and feces. D–F are the Manhattan and QQ plots of the SNPs, indels, and SV mGWAS results of ileal Staphylococcaceae. The horizontal black lines indicate significance thresholds
The co-regulation of gut microbiota and host genetics
To explore the potential correlation between host genetics and gut microbiota, we performed SNPs, indels, and SV mGWAS analysis by treating the relative abundance of families in different gut segments as phenotypes. We found that none of the three mGWAS detected significant loci for continuous traits. However, for P/A traits, SNPs, indels, and SV mGWAS detected significant loci in 54, 35, and 47 families, respectively. Among them, 15 families were detected in all three types of mGWAS (Fig. 7B, C). This suggests that the presence of different microbes in different gut segments is regulated by multiple types of genetic variations. Subsequently, we searched for families within these 15 bacterial taxa where the mGWAS results overlapped with the GWAS results for blood biochemical indicators. Unfortunately, we were unable to identify families with overlapping loci. However, it is noteworthy that among these 15 families, jejunal Veillonellaceae and Neisseriaceae showed significant positive and negative correlations with BG (r = 0.26 and − 0.21), respectively, while Staphylococcaceae in the ileum exhibited significant positive correlations with both CHOL, HDL-CH, and BG (r = 0.19, 0.23, and 0.25, Fig. 7A). We further analyzed the genetic loci of these three families and annotated the candidate genes (Supplementary Tables S15–S17). It is worth noting that for ileal Staphylococcaceae, SNP-mGWAS detected a variant located in the intergenic region between the LDAH and APOB genes at 3_102093230 (Fig. 7D–F), both of which have been extensively reported to be directly involved in cholesterol regulation. Additionally, for jejunal Veillonellaceae, SNPs and SV mGWAS identified significant loci that annotate to TOX2, and EDEM2, respectively, which are also related to blood glucose regulation (Supplementary Fig. S2A–C). For jejunal Neisseriaceae, both SNPs and SV mGWAS detected common genes in chromosome 1, but these genes are novel genes with no functional description yet (Supplementary Fig. S2D–F). Therefore, it can be inferred that there exists an interaction between Staphylococcaceae and the candidate genes LDAH and APOB in regulating cholesterol levels, and there may be a certain relationship between Veillonellaceae and TOX2 and EDEM2 in regulating blood glucose levels.
Discussion
To date, numerous studies have detected the genetic variants and genes that associated with blood biochemical indicators through various methods. These genes mainly affect the rate of metabolic pathways by influencing enzyme activity or expression, such as the involvement of hydroxy-3-methylglutaryl coenzyme A reductase (HMGCR), acetoacetyl-CoA thiolase (ACAT), and low-density lipoprotein receptor (LDLR) in cholesterol metabolism (Montoudis et al. 2008; Caselli et al. 2011; Ference et al. 2017), and the participation of insulin action and glucose uptake pathway in blood glucose metabolism (Duncan et al. 2019; Chen et al. 2022). In recent years, the impact of gut microbiota on host metabolism and phenotypic variations has been widely studied. Short-chain fatty acids (SCFAs) such as acetate, propionate, and butyrate can affect glucose and lipid metabolism when produced by gut microbiota. In addition, gut microbiota can influence inflammation and oxidative stress, which are related to the development of various blood biochemical-related diseases, such as metabolic syndrome, type 2 diabetes, cardiovascular disease, and liver diseases (Lin et al. 2012; den Besten et al. 2013; Koh et al. 2016; Chambers et al. 2018). Currently, there only exists limited research on the regulation of cholesterol, triglycerides, and blood glucose in chickens, especially with regard to multi-type genetic variants and joint regulation via microbiota in multi-gut segments. Therefore, this study conducted host whole-genome sequencing and whole-gut microbiota 16S rRNA sequencing to explore their interactive regulation on blood biochemical indicators in a population of 206 broiler chickens.
Blood biochemical indicators have a close relationship with the growth and production traits of livestock and poultry. Here, we calculated the phenotypic and genetic correlations between five blood biochemical indicators and 19 growth traits. The results showed that both CHOL and LDL-CH were negatively correlated with growth traits such as body weight, abdominal fat content, and muscle content, regardless of phenotype or genetic correlation. This indicates that an increase in the levels of these blood biochemical indicators may lead to a decrease in growth traits. Previous studies have also suggested that weight loss is commonly associated with elevated HDL-CH levels (Ginsberg 2000; Singh et al. 2007). This may be due to the augmented oxidation of fatty acid and energy expenditure that results from high cholesterol levels, ultimately leading to a decrease in body fat. In addition, glucose is also an important energy source for chicken growth, and its concentration affects the growth rate and feed efficiency of chickens (Bandyopadhyay and Das Mohapatra 2009; Hussein et al. 2018). Elevated levels of blood glucose, triglycerides, and cholesterol are associated with impaired reproductive performance and decreased milk production in dairy animals (Karcagi et al. 2010). It should be noted that the relationship between blood biochemical indicators and livestock growth traits may be influenced by various factors such as gender, age, lifestyle, and health status. Therefore, future research should further explore the complex relationships between these factors and establish more accurate models to predict their relationships.
We estimated the heritability of five blood biochemical traits based on whole-genome SNPs. Our study found that the heritability of five blood biochemical traits in chickens is low to moderate (0.15 ~ 0.24). However, there are significant differences in the heritability of blood biochemical indicators among different species, and even within the same species. For example, in humans, cholesterol has a relatively high heritability. Based on family and twin studies, the plasma level of HDL-CH appears to have a strong genetic basis, with heritability estimated at 40–60% (Lusis et al. 2004; Qasim and Rader 2006). In pigs, the heritability of cholesterol is moderate to high, ranging from 0.23 to 0.58 (Manunza et al. 2014). The heritability of serum cholesterol in chickens ranges from 0.14 to 0.57 in different studies (Dong et al. 2015; Zhang et al. 2018).
In order to capture the impact of multiple genetic variations on blood biochemical indicators, we conducted GWAS analysis based on SNPs, indels, and SVs. In GWAS analysis, the relatedness matrix is used to correct the potential confounding effect of population structure and to reduce the false positive rate. We constructed relatedness matrices based on the standardized genotype matrices of SNPs, indels, and SVs respectively, and found the relatedness matrix based on SVs exhibited significant dissimilarities with those obtained from SNPs and indels. The possible reasons include the following: (1) short-read sequencing is limited in its ability to detect large fragments of SVs (Ho et al. 2020). (2) Insufficient sequencing depth—SV detection has much higher requirements for sequencing depth than single base or small fragment variation detection. (3) The limited sample size leads to a reduced number of identified SVs.
We performed SNPs, indels, and SV GWAS analysis on five blood biochemical indicators and found the DOCK11 and RAP2C genes are the common ones detected by both SNP GWAS and indel GWAS for HDL-CH. Additionally, SV GWAS identified two genes, NTS and BOP1 in HDL-CH. DOCK11 gene functions as a guanine nucleotide exchange factor (GEF) that is dependent on MATK, a cytoplasmic tyrosine kinase (Ide et al. 2022). The DOCK11 protein promotes the GDP/GTP exchange of CDC42, thereby activating the CDC42 protein and regulating various cellular processes. Studies have shown that CDC42 controls a multi-transmembrane protein-Niemann-Pick C1-like 1 (NPC1L1) to mediate the absorption of dietary and biliary cholesterol through vesicular endocytosis. Furthermore, some studies have found that CDC42 can regulate the phosphorylation state of 3-hydroxy-3-methylglutaryl-CoA reductase (HMGCR), thereby affecting the rate of cholesterol synthesis. Then, for the RAP2C gene, it encodes proteins that belong to the Ras superfamily of small GTPases. It has been reported that overexpression of RAP2C may increase the phosphorylation level of protein kinase B (Akt), subsequently promoting the activation of mammalian target of rapamycin complex 1 (mTORC1) and leading to the activation of sterol regulatory element-binding protein (SREBP) nuclear translocation and gene transcription, thereby regulating cholesterol biosynthesis (J et al. 2018). NTS encodes a common precursor for two peptides, neuromedin N and neurotensin. Neurotensin is a neuropeptide that has been shown to be involved in the regulation of fat metabolism. Studies have demonstrated that NTS can affect insulin secretion and the metabolic activity of adipocytes, thereby influencing the overall balance of fat metabolism in the body. In addition, NTS can also regulate the release of neurotransmitters and activate the hypothalamic–pituitary–adrenal axis, further modulating fat metabolism (Feng et al. 2022). BOP1 is involved in regulating signal transduction through the p53 class mediator pathway, and p53 transcriptionally regulates squalene epoxidase to repress cholesterol synthesis (Sun et al. 2021). In terms of BG, three types of GWAS have identified three candidate genes: GDPD5, DHDH, and KCNIP1. These genes have been shown to be directly or indirectly involved in glucose regulation and are associated with insulin signaling, glucose transport, and insulin secretion. GDPD5 is a phosphodiesterase gene involved in glucose metabolism and insulin secretion regulation. DHDH is a member of the ketone metabolism pathway and is related to insulin signaling and glucose transport. The protein encoded by KCNIP1 is related to insulin secretion and insulin sensitivity. These findings contribute to a better understanding of the genetic mechanisms underlying the regulation of CHOL and BG and provide potential targets for improving the growth performance and health status of livestock and poultry.
In this study, a higher microbiability (m2) value indicated a greater influence of the gut microbiota on the host phenotype. For total CHOL, LDL-CH, and HDL-CH, cecal microbiota play more important roles than other gut segments, with m2 values of 0.20, 0.13, and 0.24, respectively. The cecum is a crucial part of the digestive tract; some bacteria presented in the cecum such as Fibrobacter, Clostridium, Odoribacter, and Akkermansia muciniphila can degrade dietary fiber, produce short-chain fatty acids, promote fat oxidation, reduce fat synthesis, and affect cholesterol metabolism (Parada Venegas et al. 2019; Cani et al. 2022). For TG and BG, the contribution of the jejunal microbiota was found to be the largest. The microbiota in the jejunum can also degrade complex carbohydrates and produce short-chain fatty acids, such as propionic, butyric, and acetic acid. Certain jejunal microbiota, such as Lactobacillus, Saccharomyces boulardii, and Butyricicoccus pullicaecorum, play an important role in glucose metabolism (Chambers et al. 2018). Moreover, certain species of Firmicutes have been shown to improve insulin sensitivity, reduce fat accumulation, and lower blood lipid levels by regulating the level of GLP-1 in the gut, thus preventing metabolic diseases such as diabetes and obesity.
The correlation and two-tailed analysis revealed an intricate connection between RF39 and Clostridia_UCG.014 with CHOL and TG. RF39 is a family within the order RF39 of the phylum Bacilli. Research on RF39 is still relatively limited, but some studies suggest that RF39 is associated with human metabolic balance. For example, a clinical study found that the abundance of RF39 was reduced in patients with type 2 diabetes (Zheng et al. 2021). Clostridia_UCG.014 is a bacterial taxon belonging to the class Clostridia in the phylum Firmicutes. Studies have shown that a decrease in the abundance of Clostridia_UCG.014 is associated with obesity (Li et al. 2021). In addition, a study has reported a positive correlation between Clostridia_UCG-014 and blood glucose levels (He et al. 2022). Streptococcaceae is a Gram-positive coccus family that includes various pathogenic bacteria such as Streptococcus pyogenes and Streptococcus pneumoniae (Wilkening and Federle 2017). However, the Streptococcus and Lactococcus genera under this family are important bacteria in industrial production that can produce lactic acid through fermentation (Kolling et al. 2012; Sahoo and Jayaraman 2019). Currently, there is limited research on the role of Streptococcaceae in human or livestock metabolism.
In order to investigate the potential relationship between host genetics and gut microbiota, we performed SNPs, indels, and SV mGWAS analysis on all families in each gut segment and found that the presence of different microbes in different gut segments is regulated by multiple types of genetic variations and the interaction between host genetics and gut microbiota. Among these families with significant loci, Staphylococcaceae in the ileum and Veillonellaceae and Neisseriaceae in the jejunum were found to be correlated with blood biochemical indicators. Staphylococcaceae is a common family of spherical bacteria, among which the only genus detected in our study, Staphylococcus, participates in important metabolic reactions such as lactate fermentation and redox reactions. Literature suggests that the abundance of Staphylococcus is positively correlated with fasting blood glucose levels, as well as 1-h and 2-h postprandial blood glucose levels (Hu et al. 2021). The key candidate genes of this family, LDAH and APOB, are important cholesterol-regulating genes. The LDAH gene encodes L-lactate dehydrogenase A-like protein, which is an NAD(H)-dependent enzyme. Studies indicate that LDAH is associated with changes in LDL cholesterol (Kory et al. 2017). The APOB gene encodes the major carrier protein, apolipoprotein B-100, of low-density lipoprotein (LDL). Apolipoprotein B-100 is the only carrier protein on LDL particles and transports cholesterol and other lipid molecules in the body (Kory et al. 2017). Our study suggests that Staphylococcaceae, LDAH, and APOB genes may interact in cholesterol utilization. Additionally, Veillonellaceae is capable of producing succinate, and an elevated level of circulating succinate is associated with impaired glucose metabolism (Serena et al. 2018). This family is associated with an important candidate gene, TOX2. TOX2 is expressed in pancreatic islet cells and is involved in regulating the synthesis and secretion of insulin, thus affecting blood glucose levels (Azarova et al. 2021). Our study suggests that the TOX2 gene may interact with Veillonellaceae in blood glucose metabolism. Neisseriaceae is a family of Gram-negative bacteria, including many bacteria that can cause human diseases, such as Neisseria gonorrhoeae and Neisseria meningitidis. This family has candidate genes such as EHBP1, CEP68, COPS9, and EED. EHBP1 (EH domain-binding protein 1) is a gene that encodes a protein involved in intracellular signaling and membrane trafficking processes. EHBP1 has been implicated in various cellular functions, including the regulation of membrane receptor signaling, the formation of membrane-bound vesicles, and the control of cell migration (Rai et al. 2020). CEP68, also known as centrosomal protein 68, is a gene that encodes a protein involved in cellular processes related to the centrosome. CEP68 is of interest in cell biology and genetics research, as its dysfunction or mutations can lead to centrosome-related abnormalities, which are associated with certain developmental disorders and diseases (Perkins et al. 2019). COPS9 is a gene that encodes a subunit of the COP9 signalosome complex. This complex is involved in the regulation of protein degradation and is associated with the ubiquitin–proteasome system (Füzesi-Levi et al. 2020). EED is a gene that encodes a protein known as Embryonic Ectoderm Development. EED is a component of the Polycomb Repressive Complex 2, which plays a crucial role in gene regulation (Cook et al. 2021). However, further research is needed to explore their interactions with Neisseriaceae.
Our study has found that blood biochemical indicators are regulated by both host genetics and gut microbiota. Multiple candidate genes were identified for the regulation of CHOL, HDL-CH, and BG. Notably, DOCK11 and RAP2C were detected in both SNPs and indels-GWAS for HDL-CH. The cecal microbiota exerted the greatest influence on cholesterol levels, whereas the jejunal microbiota had the most substantial effect on TG and BG levels. Additionally, we identified two families, RF39 and Clostridia_UCG.014 in the cecum, significantly correlated with CHOL. In addition, we also found that the presence or absence of certain families is genetically regulated and that some of these microbiotas interact with host candidate genes to regulate blood biochemical indicators, including Veillonellaceae, Neisseriaceae, and Staphylococcaceae. Our results provide novel insights into the contribution of the host genetics and gut microbiota to chicken blood biochemical indicators and may aid in developing strategies to manipulate blood biochemical indicators and their associated growth traits.
Data availability
The raw data are available from the Sequence Read Archive with accession numbers PRJNA449436, PRJNA449437, and PRJNA449438.
References
Alonge M, Wang X, Benoit M, Soyk S, Pereira L, Zhang L, Suresh H, Ramakrishnan S, Maumus F, Ciren D, Levy Y, Harel TH, Shalev-Schlosser G, Amsellem Z, Razifard H, Caicedo AL, Tieman DM, Klee H, Kirsche M, Aganezov S, Ranallo-Benavidez TR, Lemmon ZH, Kim J, Robitaille G, Kramer M, Goodwin S, McCombie WR, Hutton S, Van Eck J, Gillis J, Eshed Y, Sedlazeck FJ, van der Knaap E, Schatz MC, Lippman ZB (2020) Major impacts of widespread structural variation on gene expression and crop improvement in tomato. Cell 182:145-161.e23. https://doi.org/10.1016/j.cell.2020.05.021
Arsenault BJ, Lemieux I, Després J-P, Wareham NJ, Kastelein JJP, Khaw K-T, Boekholdt SM (2010) The hypertriglyceridemic-waist phenotype and the risk of coronary artery disease: results from the EPIC-Norfolk prospective population study. CMAJ 182:1427–1432. https://doi.org/10.1503/cmaj.091276
Azarova I, Klyosova E, Polonikov A (2021) The link between type 2 diabetes mellitus and the polymorphisms of glutathione-metabolizing genes suggests a new hypothesis explaining disease initiation and progression. Life (basel) 11:886. https://doi.org/10.3390/life11090886
Bandyopadhyay P, Das Mohapatra PK (2009) Effect of a probiotic bacterium Bacillus circulans PB7 in the formulated diets: on growth, nutritional quality and immunity of Catla catla (Ham.). Fish Physiol Biochem 35:467–478. https://doi.org/10.1007/s10695-008-9272-8
Biscay Lirio R, Valdés Sosa PA, Pascual Marqui RD, Jiménez-Sobrino JC, Alvarez Amador A, Galán Garcia L (1989) Multivariate Box-Cox transformations with applications to neurometric data. Comput Biol Med 19:263–267. https://doi.org/10.1016/0010-4825(89)90013-9
Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, Bai Y, Bisanz JE, Bittinger K, Brejnrod A, Brislawn CJ, Brown CT, Callahan BJ, Caraballo-Rodríguez AM, Chase J, Cope EK, Da Silva R, Diener C, Dorrestein PC, Douglas GM, Durall DM, Duvallet C, Edwardson CF, Ernst M, Estaki M, Fouquier J, Gauglitz JM, Gibbons SM, Gibson DL, Gonzalez A, Gorlick K, Guo J, Hillmann B, Holmes S, Holste H, Huttenhower C, Huttley GA, Janssen S, Jarmusch AK, Jiang L, Kaehler BD, Kang KB, Keefe CR, Keim P, Kelley ST, Knights D, Koester I, Kosciolek T, Kreps J, Langille MGI, Lee J, Ley R, Liu Y-X, Loftfield E, Lozupone C, Maher M, Marotz C, Martin BD, McDonald D, McIver LJ, Melnik AV, Metcalf JL, Morgan SC, Morton JT, Naimey AT, Navas-Molina JA, Nothias LF, Orchanian SB, Pearson T, Peoples SL, Petras D, Preuss ML, Pruesse E, Rasmussen LB, Rivers A, Robeson MS, Rosenthal P, Segata N, Shaffer M, Shiffer A, Sinha R, Song SJ, Spear JR, Swafford AD, Thompson LR, Torres PJ, Trinh P, Tripathi A, Turnbaugh PJ, Ul-Hasan S, vander Hooft JJJ, Vargas F, Vázquez-Baeza Y, Vogtmann E, von Hippel M, Walters W, Wan Y, Wang M, Warren J, Weber KC, Williamson CHD, Willis AD, Xu ZZ, Zaneveld JR, Zhang Y, Zhu Q, Knight R, Caporaso JG (2019) Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 37:852–857. https://doi.org/10.1038/s41587-019-0209-9
Browning SR, Browning BL (2007) Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet 81:1084–1097. https://doi.org/10.1086/521987
Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP (2016) DADA2: high resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583. https://doi.org/10.1038/nmeth.3869
Callahan BJ, McMurdie PJ, Holmes SP (2017) Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J 11:2639–2643. https://doi.org/10.1038/ismej.2017.119
Cani PD, Depommier C, Derrien M, Everard A, de Vos WM (2022) Akkermansia muciniphila: paradigm for next-generation beneficial microorganisms. Nat Rev Gastroenterol Hepatol 19:625–637. https://doi.org/10.1038/s41575-022-00631-9
Caselli RJ, Dueck AC, Locke DEC, Sabbagh MN, Ahern GL, Rapcsak SZ, Baxter LC, Yaari R, Woodruff BK, Hoffman-Snyder C, Rademakers R, Findley S, Reiman EM (2011) Cerebrovascular risk factors and preclinical memory decline in healthy APOE ε4 homozygotes. Neurology 76:1078–1084. https://doi.org/10.1212/WNL.0b013e318211c3ae
Chambers ES, Preston T, Frost G, Morrison DJ (2018) Role of gut microbiota-generated short-chain fatty acids in metabolic and cardiovascular health. Curr Nutr Rep 7:198–206. https://doi.org/10.1007/s13668-018-0248-8
Chen X, Schulz-Trieglaff O, Shaw R, Barnes B, Schlesinger F, Källberg M, Cox AJ, Kruglyak S, Saunders CT (2016) Manta: rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics 32:1220–1222. https://doi.org/10.1093/bioinformatics/btv710
Chen B, Du Y-R, Zhu H, Sun M-L, Wang C, Cheng Y, Pang H, Ding G, Gao J, Tan Y, Tong X, Lv P, Zhou F, Zhan Q, Xu Z-M, Wang L, Luo D, Ye Y, Jin L, Zhang S, Zhu Y, Lin X, Wu Y, Jin L, Zhou Y, Yan C, Sheng J, Flatt PR, Xu G-L, Huang H (2022) Maternal inheritance of glucose intolerance via oocyte TET3 insufficiency. Nature 605:761–766. https://doi.org/10.1038/s41586-022-04756-4
Choe JH, Kim BC (2014) Association of blood glucose, blood lactate, serum cortisol levels, muscle metabolites, muscle fiber type composition, and pork quality traits. Meat Sci 97:137–142. https://doi.org/10.1016/j.meatsci.2014.01.024
Clevenger J, Chavarro C, Pearl SA, Ozias-Akins P, Jackson SA (2015) Single nucleotide polymorphism identification in polyploids: a review, example, and recommendations. Mol Plant 8:831–846. https://doi.org/10.1016/j.molp.2015.02.002
Cohen J, Pertsemlidis A, Kotowski IK, Graham R, Garcia CK, Hobbs HH (2005) Low LDL cholesterol in individuals of African descent resulting from frequent nonsense mutations in PCSK9. Nat Genet 37:161–165. https://doi.org/10.1038/ng1509
Cook N, Chen J, Zhou J, Wu D (2021) Embryonic ectoderm development (EED) as a novel target for cancer treatment. Curr Top Med Chem 21:2771–2777. https://doi.org/10.2174/1568026621666210920154942
Danecek P, Bonfield JK, Liddle J, Marshall J, Ohan V, Pollard MO, Whitwham A, Keane T, McCarthy SA, Davies RM, Li H (2021) Twelve years of SAMtools and BCFtools. Gigascience 10:giab008. https://doi.org/10.1093/gigascience/giab008
Deary IJ, Yang J, Davies G, Harris SE, Tenesa A, Liewald D, Luciano M, Lopez LM, Gow AJ, Corley J, Redmond P, Fox HC, Rowe SJ, Haggarty P, McNeill G, Goddard ME, Porteous DJ, Whalley LJ, Starr JM, Visscher PM (2012) Genetic contributions to stability and change in intelligence from childhood to old age. Nature 482:212–215. https://doi.org/10.1038/nature10781
den Besten G, van Eunen K, Groen AK, Venema K, Reijngoud D-J, Bakker BM (2013) The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J Lipid Res 54:2325–2340. https://doi.org/10.1194/jlr.R036012
Dong J-Q, Zhang H, Jiang X-F, Wang S-Z, Du Z-Q, Wang Z-P, Leng L, Cao Z-P, Li Y-M, Luan P, Li H (2015) Comparison of serum biochemical parameters between two broiler chicken lines divergently selected for abdominal fat content. J Anim Sci 93:3278–3286. https://doi.org/10.2527/jas.2015-8871
Duncan A, Heyer MP, Ishikawa M, Caligiuri SPB, Liu X-A, Chen Z, Micioni Di Bonaventura MV, Elayouby KS, Ables JL, Howe WM, Bali P, Fillinger C, Williams M, O’Connor RM, Wang Z, Lu Q, Kamenecka TM, Ma’ayan A, O’Neill HC, Ibanez-Tallon I, Geurts AM, Kenny PJ (2019) Habenular TCF7L2 links nicotine addiction to diabetes. Nature 574:372–377. https://doi.org/10.1038/s41586-019-1653-x
Feng X, Zhang Y, Du M, Li S, Ding J, Wang J, Wang Y, Liu P (2022) Identification of diagnostic biomarkers and therapeutic targets in peripheral immune landscape from coronary artery disease. J Transl Med 20:399. https://doi.org/10.1186/s12967-022-03614-1
Ference BA, Kastelein JJP, Ginsberg HN, Chapman MJ, Nicholls SJ, Ray KK, Packard CJ, Laufs U, Brook RD, Oliver-Williams C, Butterworth AS, Danesh J, Smith GD, Catapano AL, Sabatine MS (2017) Association of genetic variants related to CETP Inhibitors and statins with lipoprotein levels and cardiovascular risk. JAMA 318:947–956. https://doi.org/10.1001/jama.2017.11467
Füzesi-Levi MG, Fainer I, Ivanov Enchev R, Ben-Nissan G, Levin Y, Kupervaser M, Friedlander G, Salame TM, Nevo R, Peter M, Sharon M (2020) CSNAP, the smallest CSN subunit, modulates proteostasis through cullin-RING ubiquitin ligases. Cell Death Differ 27:984–998. https://doi.org/10.1038/s41418-019-0392-8
Gacesa R, Kurilshikov A, Vich Vila A, Sinha T, Klaassen MAY, Bolte LA, Andreu-Sánchez S, Chen L, Collij V, Hu S, Dekens JAM, Lenters VC, Björk JR, Swarte JC, Swertz MA, Jansen BH, Gelderloos-Arends J, Jankipersadsing S, Hofker M, Vermeulen RCH, Sanna S, Harmsen HJM, Wijmenga C, Fu J, Zhernakova A, Weersma RK (2022) Environmental factors shaping the gut microbiome in a Dutch population. Nature 604:732–739. https://doi.org/10.1038/s41586-022-04567-7
Ginsberg HN (2000) Nonpharmacologic management of low levels of high-density lipoprotein cholesterol. Am J Cardiol 86(12A):41L-45L. https://doi.org/10.1016/s0002-9149(00)01469-7
Grieneisen L, Dasari M, Gould TJ, Björk JR, Grenier J-C, Yotova V, Jansen D, Gottel N, Gordon JB, Learn NH, Gesquiere LR, Wango TL, Mututua RS, Warutere JK, Siodi L, Gilbert JA, Barreiro LB, Alberts SC, Tung J, Archie EA, Blekhman R (2021) Gut microbiome heritability is nearly universal but environmentally contingent. Science 373:181–186. https://doi.org/10.1126/science.aba5483
He G, Chen T, Huang L, Zhang Y, Feng Y, Qu S, Yin X, Liang L, Yan J, Liu W (2022) Tremella fuciformis polysaccharide reduces obesity in high-fat diet-fed mice by modulation of gut microbiota. Front Microbiol 13:1073350. https://doi.org/10.3389/fmicb.2022.1073350
Heianza Y, Sun D, Li X, DiDonato JA, Bray GA, Sacks FM, Qi L (2019) Gut microbiota metabolites, amino acid metabolites and improvements in insulin sensitivity and glucose metabolism: the POUNDS Lost trial. Gut 68:263–270. https://doi.org/10.1136/gutjnl-2018-316155
Hermier D, Chapman MJ, Leclercq B (1984) Plasma lipoprotein profile in fasted and refed chickens of two strains selected for high or low adiposity. J Nutr 114:1112–1121. https://doi.org/10.1093/jn/114.6.1112
Hernandez-Segura A, Rubingh R, Demaria M (2019) Identification of stable senescence-associated reference genes. Aging Cell 18:e12911. https://doi.org/10.1111/acel.12911
Ho SS, Urban AE, Mills RE (2020) Structural variation in the sequencing era. Nat Rev Genet 21:171–189. https://doi.org/10.1038/s41576-019-0180-9
Hofmann AL, Behr J, Singer J, Kuipers J, Beisel C, Schraml P, Moch H, Beerenwinkel N (2017) Detailed simulation of cancer exome sequencing data reveals differences and common limitations of variant callers. BMC Bioinf 18:8. https://doi.org/10.1186/s12859-016-1417-7
Hu P, Chen X, Chu X, Fan M, Ye Y, Wang Y, Han M, Yang X, Yuan J, Zha L, Zhao B, Yang C-X, Qi X-R, Ning K, Debelius J, Ye W, Xiong B, Pan X-F, Pan A (2021) Association of gut microbiota during early pregnancy with risk of incident gestational diabetes mellitus. J Clin Endocrinol Metab 106:e4128–e4141. https://doi.org/10.1210/clinem/dgab346
Hu H, Shao W, Liu Q, Liu N, Wang Q, Xu J, Zhang X, Weng Z, Lu Q, Jiao L, Chen C, Sun H, Jiang Z, Zhang X, Gu A (2022) Gut microbiota promotes cholesterol gallstone formation by modulating bile acid composition and biliary cholesterol secretion. Nat Commun 13:252. https://doi.org/10.1038/s41467-021-27758-8
Hughes DA (2020) Genome-wide associations of human gut microbiome variation and implications for causal inference analyses. Nature Microbiol 5:1079–1087. https://doi.org/10.1038/s41564-020-0743-8
Hussein AS, Ayoub MA, Elhwetiy AY, Ghurair JA, Sulaiman M, Habib HM (2018) Effect of dietary inclusion of sugar syrup on production performance, egg quality and blood biochemical parameters in laying hens. Anim Nutr 4:59–64. https://doi.org/10.1016/j.aninu.2017.11.001
Ide M, Tabata N, Yonemura Y, Shirasaki T, Murai K, Wang Y, Ishida A, Okada H, Honda M, Kaneko S, Doi N, Ito S, Yanagawa H (2022) Guanine nucleotide exchange factor DOCK11-binding peptide fused with a single chain antibody inhibits hepatitis B virus infection and replication. J Biol Chem 298:102097. https://doi.org/10.1016/j.jbc.2022.102097
Jeffares DC, Jolly C, Hoti M, Speed D, Shaw L, Rallis C, Balloux F, Dessimoz C, Bähler J, Sedlazeck FJ (2017) Transient structural variations have strong effects on quantitative traits and reproductive isolation in fission yeast. Nat Commun 8:14061. https://doi.org/10.1038/ncomms14061
Jiang Z, Cherian G, Robinson FE, Sim JS (1990) Effect of feeding cholesterol to laying hens and chicks on cholesterol metabolism in pre- and posthatch chicks. Poult Sci 69:1694–1701. https://doi.org/10.3382/ps.0691694
Karcagi RG, Gaál T, Ribiczey P, Huszenicza G, Husvéth F (2010) Milk production, peripartal liver triglyceride concentration and plasma metabolites of dairy cows fed diets supplemented with calcium soaps or hydrogenated triglycerides of palm oil. J Dairy Res 77:151–158. https://doi.org/10.1017/S0022029909990604
Karlsson FH, Tremaroli V, Nookaew I, Bergström G, Behre CJ, Fagerberg B, Nielsen J, Bäckhed F (2013) Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature 498:99–103. https://doi.org/10.1038/nature12198
Kathiresan S, Willer CJ, Peloso GM, Demissie S, Musunuru K, Schadt EE, Kaplan L, Bennett D, Li Y, Tanaka T, Voight BF, Bonnycastle LL, Jackson AU, Crawford G, Surti A, Guiducci C, Burtt NP, Parish S, Clarke R, Zelenika D, Kubalanza KA, Morken MA, Scott LJ, Stringham HM, Galan P, Swift AJ, Kuusisto J, Bergman RN, Sundvall J, Laakso M, Ferrucci L, Scheet P, Sanna S, Uda M, Yang Q, Lunetta KL, Dupuis J, de Bakker PIW, O’Donnell CJ, Chambers JC, Kooner JS, Hercberg S, Meneton P, Lakatta EG, Scuteri A, Schlessinger D, Tuomilehto J, Collins FS, Groop L, Altshuler D, Collins R, Lathrop GM, Melander O, Salomaa V, Peltonen L, Orho-Melander M, Ordovas JM, Boehnke M, Abecasis GR, Mohlke KL, Cupples LA (2009) Common variants at 30 loci contribute to polygenic dyslipidemia. Nat Genet 41:56–65. https://doi.org/10.1038/ng.291
Koh A, De Vadder F, Kovatcheva-Datchary P, Bäckhed F (2016) From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell 165:1332–1345. https://doi.org/10.1016/j.cell.2016.05.041
Kolling GL, Wu M, Warren CA, Durmaz E, Klaenhammer TR, Timko MP, Guerrant RL (2012) Lactic acid production by Streptococcus thermophilus alters Clostridium difficile infection and in vitro Toxin A production. Gut Microbes 3:523–529. https://doi.org/10.4161/gmic.21757
Kory N, Grond S, Kamat SS, Li Z, Krahmer N, Chitraju C, Zhou P, Fröhlich F, Semova I, Ejsing C, Zechner R, Cravatt BF, Farese RV, Walther TC (2017) Mice lacking lipid droplet-associated hydrolase, a gene linked to human prostate cancer, have normal cholesterol ester metabolism. J Lipid Res 58:226–235. https://doi.org/10.1194/jlr.M072538
Li Z, Votava JA, Zajac GJM, Nguyen JN, Leyva Jaimes FB, Ly SM, Brinkman JA, De Giorgi M, Kaul S, Green CL, St Clair SL, Belisle SL, Rios JM, Nelson DW, Sorci-Thomas MG, Lagor WR, Lamming DW, Eric Yen C-L, Parks BW (2020) Integrating mouse and human genetic data to move beyond GWAS and identify causal genes in cholesterol metabolism. Cell Metab 31:741-754.e5. https://doi.org/10.1016/j.cmet.2020.02.015
Li K, Epperly MW, Barreto GA, Greenberger JS, Methé BA (2021) Longitudinal fecal microbiome study of total body irradiated mice treated with radiation mitigators identifies bacterial associations with survival. Front Cell Infect Microbiol 11:715396. https://doi.org/10.3389/fcimb.2021.715396
Lin HV, Frassetto A, Kowalik EJ, Nawrocki AR, Lu MM, Kosinski JR, Hubert JA, Szeto D, Yao X, Forrest G, Marsh DJ (2012) Butyrate and propionate protect against diet-induced obesity and regulate gut hormones via free fatty acid receptor 3-independent mechanisms. PLoS One 7:e35240. https://doi.org/10.1371/journal.pone.0035240
Luo J, Yang H, Song B-L (2020) Mechanisms and regulation of cholesterol homeostasis. Nat Rev Mol Cell Biol 21:225–245. https://doi.org/10.1038/s41580-019-0190-7
Lusis AJ, Mar R, Pajukanta P (2004) Genetics of atherosclerosis. Annu Rev Genomics Hum Genet 5:189–218. https://doi.org/10.1146/annurev.genom.5.061903.175930
Mahajan A, Jankovic J, Marsh L, Patel A, Jinnah HA, Comella C, Barbano R, Perlmutter J, Patel N, members of the Dystonia Coalition (2018) Cervical dystonia and substance abuse. J Neurol 265:970–975. https://doi.org/10.1007/s00415-018-8840-9
Manunza A, Casellas J, Quintanilla R, González-Prendes R, Pena RN, Tibau J, Mercadé A, Castelló A, Aznárez N, Hernández-Sánchez J, Amills M (2014) A genome-wide association analysis for porcine serum lipid traits reveals the existence of age-specific genetic determinants. BMC Genomics 15:758. https://doi.org/10.1186/1471-2164-15-758
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA (2010) The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303. https://doi.org/10.1101/gr.107524.110
Montoudis A, Seidman E, Boudreau F, Beaulieu J-F, Menard D, Elchebly M, Mailhot G, Sane A-T, Lambert M, Delvin E, Levy E (2008) Intestinal fatty acid binding protein regulates mitochondrion beta-oxidation and cholesterol uptake. J Lipid Res 49:961–972. https://doi.org/10.1194/jlr.M700363-JLR200
Parada Venegas D, De la Fuente MK, Landskron G, González MJ, Quera R, Dijkstra G, Harmsen HJM, Faber KN, Hermoso MA (2019) Short chain fatty acids (SCFAs)-mediated gut epithelial and immune regulation and its relevance for inflammatory bowel diseases. Front Immunol 10:277. https://doi.org/10.3389/fimmu.2019.00277
Perkins JR, Acosta-Herrera M, Plaza-Serón MC, Jurado-Escobar R, Doña I, García-Martín E, Isidoro-García M, Bartra J, Ribas-Perez D, Mayorga C, Torres MJ, Flores C, Cornejo-García JA (2019) Polymorphisms in CEP68 gene associated with risk of immediate selective reactions to non-steroidal anti-inflammatory drugs. Pharmacogenomics J 19:191–199. https://doi.org/10.1038/s41397-018-0038-0
Price AL, Zaitlen NA, Reich D, Patterson N (2010) New approaches to population stratification in genome-wide association studies. Nat Rev Genet 11:459–463. https://doi.org/10.1038/nrg2813
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, Sham PC (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81:559–575. https://doi.org/10.1086/519795
Qasim A, Rader DJ (2006) Human genetics of variation in high-density lipoprotein cholesterol. Curr Artherosclr Rep 8:198–205. https://doi.org/10.1007/s11883-006-0074-0
Qin P, Lu H, Du H, Wang H, Chen W, Chen Z, He Q, Ou S, Zhang H, Li X, Li X, Li Y, Liao Y, Gao Q, Tu B, Yuan H, Ma B, Wang Y, Qian Y, Fan S, Li W, Wang J, He M, Yin J, Li T, Jiang N, Chen X, Liang C, Li S (2021) Pan-genome analysis of 33 genetically diverse rice accessions reveals hidden genomic variations. Cell 184:3542-3558.e16. https://doi.org/10.1016/j.cell.2021.04.046
Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO (2013) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590-596. https://doi.org/10.1093/nar/gks1219
Rai A, Bleimling N, Vetter IR, Goody RS (2020) The mechanism of activation of the actin binding protein EHBP1 by Rab8 family members. Nat Commun 11:4187. https://doi.org/10.1038/s41467-020-17792-3
Rausch T, Zichner T, Schlattl A, Stutz AM, Benes V, Korbel JO (2012) DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics 28:i333–i339. https://doi.org/10.1093/bioinformatics/bts378
Sahoo TK, Jayaraman G (2019) Co-culture of Lactobacillus delbrueckii and engineered Lactococcus lactis enhances stoichiometric yield of D-lactic acid from whey permeate. Appl Microbiol Biotechnol 103:5653–5662. https://doi.org/10.1007/s00253-019-09819-7
Sedgwick P (2014) Multiple hypothesis testing and Bonferroni’s correction. BMJ 349:g6284. https://doi.org/10.1136/bmj.g6284
Serena C, Ceperuelo-Mallafré V, Keiran N, Queipo-Ortuño MI, Bernal R, Gomez-Huelgas R, Urpi-Sarda M, Sabater M, Pérez-Brocal V, Andrés-Lacueva C, Moya A, Tinahones FJ, Fernández-Real JM, Vendrell J, Fernández-Veledo S (2018) Elevated circulating levels of succinate in human obesity are linked to specific gut microbiota. ISME J 12:1642–1657. https://doi.org/10.1038/s41396-018-0068-2
Singh IM, Shishehbor MH, Ansell BJ (2007) High-density lipoprotein as a therapeutic target: a systematic review. JAMA 298(7):786–798. https://doi.org/10.1001/jama.298.7.786
Sun H, Li L, Li W, Yang F, Zhang Z, Liu Z, Du W (2021) p53 transcriptionally regulates SQLE to repress cholesterol synthesis and tumor growth. EMBO Rep 22:e52537. https://doi.org/10.15252/embr.202152537
Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, Pirruccello JP, Ripatti S, Chasman DI, Willer CJ, Johansen CT, Fouchier SW, Isaacs A, Peloso GM, Barbalic M, Ricketts SL, Bis JC, Aulchenko YS, Thorleifsson G, Feitosa MF, Chambers J, Orho-Melander M, Melander O, Johnson T, Li X, Guo X, Li M, Shin Cho Y, Jin Go M, Jin Kim Y, Lee J-Y, Park T, Kim K, Sim X, Twee-Hee Ong R, Croteau-Chonka DC, Lange LA, Smith JD, Song K, Hua Zhao J, Yuan X, Luan J, Lamina C, Ziegler A, Zhang W, Zee RYL, Wright AF, Witteman JCM, Wilson JF, Willemsen G, Wichmann H-E, Whitfield JB, Waterworth DM, Wareham NJ, Waeber G, Vollenweider P, Voight BF, Vitart V, Uitterlinden AG, Uda M, Tuomilehto J, Thompson JR, Tanaka T, Surakka I, Stringham HM, Spector TD, Soranzo N, Smit JH, Sinisalo J, Silander K, Sijbrands EJG, Scuteri A, Scott J, Schlessinger D, Sanna S, Salomaa V, Saharinen J, Sabatti C, Ruokonen A, Rudan I, Rose LM, Roberts R, Rieder M, Psaty BM, Pramstaller PP, Pichler I, Perola M, Penninx BWJH, Pedersen NL, Pattaro C, Parker AN, Pare G, Oostra BA, O’Donnell CJ, Nieminen MS, Nickerson DA, Montgomery GW, Meitinger T, McPherson R, McCarthy MI, McArdle W, Masson D, Martin NG, Marroni F, Mangino M, Magnusson PKE, Lucas G, Luben R, Loos RJF, Lokki M-L, Lettre G, Langenberg C, Launer LJ, Lakatta EG, Laaksonen R, Kyvik KO, Kronenberg F, König IR, Khaw K-T, Kaprio J, Kaplan LM, Johansson A, Jarvelin M-R, Janssens ACJW, Ingelsson E, Igl W, Kees Hovingh G, Hottenga J-J, Hofman A, Hicks AA, Hengstenberg C, Heid IM, Hayward C, Havulinna AS, Hastie ND, Harris TB, Haritunians T, Hall AS, Gyllensten U, Guiducci C, Groop LC, Gonzalez E, Gieger C, Freimer NB, Ferrucci L, Erdmann J, Elliott P, Ejebe KG, Döring A, Dominiczak AF, Demissie S, Deloukas P, de Geus EJC, de Faire U, Crawford G, Collins FS, Chen YI, Caulfield MJ, Campbell H, Burtt NP, Bonnycastle LL, Boomsma DI, Boekholdt SM, Bergman RN, Barroso I, Bandinelli S, Ballantyne CM, Assimes TL, Quertermous T, Altshuler D, Seielstad M, Wong TY, Tai E-S, Feranil AB, Kuzawa CW, Adair LS, Taylor HA, Borecki IB, Gabriel SB, Wilson JG, Holm H, Thorsteinsdottir U, Gudnason V, Krauss RM, Mohlke KL, Ordovas JM, Munroe PB, Kooner JS, Tall AR, Hegele RA, Kastelein JJP, Schadt EE, Rotter JI, Boerwinkle E, Strachan DP, Mooser V, Stefansson K, Reilly MP, Samani NJ, Schunkert H, Cupples LA, Sandhu MS, Ridker PM, Rader DJ, van Duijn CM, Peltonen L, Abecasis GR, Boehnke M, Kathiresan S (2010) Biological, clinical and population relevance of 95 loci for blood lipids. Nature 466:707–713. https://doi.org/10.1038/nature09270
Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI (2006) An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444:1027–1031. https://doi.org/10.1038/nature05414
Voight BF, Peloso GM, Orho-Melander M, Frikke-Schmidt R, Barbalic M, Jensen MK, Hindy G, Hólm H, Ding EL, Johnson T, Schunkert H, Samani NJ, Clarke R, Hopewell JC, Thompson JF, Li M, Thorleifsson G, Newton-Cheh C, Musunuru K, Pirruccello JP, Saleheen D, Chen L, Stewart AFR, Schillert A, Thorsteinsdottir U, Thorgeirsson G, Anand S, Engert JC, Morgan T, Spertus J, Stoll M, Berger K, Martinelli N, Girelli D, McKeown PP, Patterson CC, Epstein SE, Devaney J, Burnett M-S, Mooser V, Ripatti S, Surakka I, Nieminen MS, Sinisalo J, Lokki M-L, Perola M, Havulinna A, de Faire U, Gigante B, Ingelsson E, Zeller T, Wild P, de Bakker PIW, Klungel OH, Maitland-van der Zee A-H, Peters BJM, de Boer A, Grobbee DE, Kamphuisen PW, Deneer VHM, Elbers CC, Onland-Moret NC, Hofker MH, Wijmenga C, Verschuren WMM, Boer JMA, van der Schouw YT, Rasheed A, Frossard P, Demissie S, Willer C, Do R, Ordovas JM, Abecasis GR, Boehnke M, Mohlke KL, Daly MJ, Guiducci C, Burtt NP, Surti A, Gonzalez E, Purcell S, Gabriel S, Marrugat J, Peden J, Erdmann J, Diemert P, Willenborg C, König IR, Fischer M, Hengstenberg C, Ziegler A, Buysschaert I, Lambrechts D, Van de Werf F, Fox KA, El Mokhtari NE, Rubin D, Schrezenmeir J, Schreiber S, Schäfer A, Danesh J, Blankenberg S, Roberts R, McPherson R, Watkins H, Hall AS, Overvad K, Rimm E, Boerwinkle E, Tybjaerg-Hansen A, Cupples LA, Reilly MP, Melander O, Mannucci PM, Ardissino D, Siscovick D, Elosua R, Stefansson K, O’Donnell CJ, Salomaa V, Rader DJ, Peltonen L, Schwartz SM, Altshuler D, Kathiresan S (2012) Plasma HDL cholesterol and risk of myocardial infarction: a mendelian randomisation study. Lancet 380:572–580. https://doi.org/10.1016/S0140-6736(12)60312-2
Wan Y, Dong P, Zhu X, Lei Y, Shen J, Liu W, Liu K, Zhang X (2022) Bibliometric and visual analysis of intestinal ischemia reperfusion from 2004 to 2022. Front Med (lausanne) 9:963104. https://doi.org/10.3389/fmed.2022.963104
Wang K, Li M, Hakonarson H (2010) ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38:e164. https://doi.org/10.1093/nar/gkq603
Wen C, Yan W, Sun C, Ji C, Zhou Q, Zhang D, Zheng J, Yang N (2019) The gut microbiota is largely independent of host genetics in regulating fat deposition in chickens. ISME J 13:1422–1436. https://doi.org/10.1038/s41396-019-0367-2
Wen C, Yan W, Mai C, Duan Z, Zheng J, Sun C, Yang N (2021) Joint contributions of the gut microbiota and host genetics to feed efficiency in chickens. Microbiome 9:126. https://doi.org/10.1186/s40168-021-01040-x
Wilkening RV, Federle MJ (2017) Evolutionary constraints shaping Streptococcus pyogenes-host interactions. Trends Microbiol 25:562–572. https://doi.org/10.1016/j.tim.2017.01.007
Yan W, Sun C, Wen C, Ji C, Zhang D, Yang N (2019) Relationships between feeding behaviors and performance traits in slow-growing yellow broilers. Poult Sci 98:548–555. https://doi.org/10.3382/ps/pey424
Yang J, Lee SH, Goddard ME, Visscher PM (2011) GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet 88:76–82. https://doi.org/10.1016/j.ajhg.2010.11.011
Yang H-L, Kong L, Hou L-L, Shen H-F, Wang Y, Gu X-G, Qin J-M, Yin P-H, Li Q (2012) Analysis of risk factors for polypoid lesions of gallbladder among health examinees. World J Gastroenterol 18:3015–3019. https://doi.org/10.3748/wjg.v18.i23.3015
Yang T, Santisteban MM, Rodriguez V, Li E, Ahmari N, Carvajal JM, Zadeh M, Gong M, Qi Y, Zubcevic J, Sahay B, Pepine CJ, Raizada MK, Mohamadzadeh M (2015) Gut dysbiosis is linked to hypertension. Hypertension 65:1331–1340. https://doi.org/10.1161/HYPERTENSIONAHA.115.05315
Yen JL, Garcia S, Montana A, Harris J, Chervitz S, Morra M, West J, Chen R, Church DM (2017) A variant by any name: quantifying annotation discordance across tools and clinical databases. Genome Med 9:7. https://doi.org/10.1186/s13073-016-0396-7
Zhang HL, Xu ZQ, Yang LL, Wang YX, Li YM, Dong JQ, Zhang XY, Jiang XY, Jiang XF, Li H, Zhang DX, Zhang H (2018) Genetic parameters for the prediction of abdominal fat traits using blood biochemical indicators in broilers. Br Poult Sci 59:28–33. https://doi.org/10.1080/00071668.2017.1379052
Zheng H, Xu P, Jiang Q, Xu Q, Zheng Y, Yan J, Ji H, Ning J, Zhang X, Li C, Zhang L, Li Y, Li X, Song W, Gao H (2021) Depletion of acetate-producing bacteria from the gut microbiota facilitates cognitive impairment through the gut-brain neural mechanism in diabetic mice. Microbiome 9:145. https://doi.org/10.1186/s40168-021-01088-9
Zhou X, Stephens M (2012) Genome-wide efficient mixed-model analysis for association studies. Nat Genet 44:821–824. https://doi.org/10.1038/ng.2310
Funding
This study was supported by the National Natural Science Foundation of China (32102535 and 31930105) and China Agriculture Research Systems (CARS-40).
Author information
Authors and Affiliations
Contributions
C. S., C. W., and N. Y. designed the study. X. J. performed the analysis and wrote the manuscript. B. Z., F. L., C. Z., and J. J. assisted in data analyzing. C. S. and C. W. contributed to the revisions. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval
The protocol was approved by the Animal Care and Use Committee of China Agricultural University (Permit Number: AW08059102–1).
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Jiang, X., Zhang, B., Lan, F. et al. Host genetics and gut microbiota jointly regulate blood biochemical indicators in chickens. Appl Microbiol Biotechnol 107, 7601–7620 (2023). https://doi.org/10.1007/s00253-023-12814-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-023-12814-8