Abstract
The carboxylic acid propionate is a valuable platform chemical with applications in various fields. The biological production of this acid has become of great interest as it can be considered a sustainable alternative to petrochemical synthesis. In this work, Clostridium saccharoperbutylacetonicum was metabolically engineered to produce propionate via the acrylate pathway. In total, the established synthetic pathway comprised eight genes encoding the enzymes catalyzing the conversion of pyruvate to propionate. These included the propionate CoA-transferase, the lactoyl-CoA dehydratase, and the acryloyl-CoA reductase from Anaerotignum neopropionicum as well as a D-lactate dehydrogenase from Leuconostoc mesenteroides subsp. mesenteroides. Due to difficulties in assembling all genes on one plasmid under the control of standard promoters, the PtcdB-tcdR promoter system from Clostridium difficile was integrated into a two-plasmid system carrying the acrylate pathway genes. Several promoters were analyzed for their activity in C. saccharoperbutylacetonicum using the fluorescence-activating and absorption-shifting tag (FAST) as a fluorescent reporter to identify suitable candidates to drive tcdR expression. After selecting the lactose-inducible PbgaL promoter, engineered C. saccharoperbutylacetonicum strains produced 0.7 mM propionate upon induction of gene expression. The low productivity was suspected to be a consequence of a metabolic imbalance leading to acryloyl-CoA accumulation in the cells. To even out the proposed imbalance, the propionate-synthesis operons were rearranged, thereby increasing the propionate concentration by almost four-fold. This study is the first one to report recombinant propionate production using a clostridial host strain that has opened a new path towards bio-based propionate to be improved further in subsequent work.
Key points
• Determination of promoter activities in C. saccharoperbutylacetonicum using FAST.
• Implementation of propionate production in C. saccharoperbutylacetonicum.
• Elevation of propionate production by 375% to a concentration of 3 mM.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Propionate is a valuable platform chemical with a wide range of applications. Due to its antimicrobial activity, it is mostly used as a food and feed preservative, an ingredient in cleaning agents, or as an herbicide. It also gains increasing importance in the production of pharmaceuticals, plastics, and cosmetics (Gonzalez-Garcia et al. 2017; Samel et al. 2018). Moreover, it is considered an important precursor chemical as it is often esterified with short-chain alcohols, olefins, or acetylenes to yield corresponding alcohol or vinyl esters, which themselves have versatile applications (Samel et al. 2018). Propionate synthesis is currently achieved via chemical processes, i.e., the carbonylation of ethylene or the oxidation of propionaldehyde (Samel et al. 2018). However, bio-based approaches using bacteria as cell factories for propionate production from cheap or waste-derived substrates become increasingly attractive as sustainable alternatives to petrochemical production (Stowers et al. 2014). Although not commercially profitable yet, the desire to achieve an environmentally friendly propionate production has led to multiple studies exploring the capabilities of bacteria in that regard. There are different bacterial species that can naturally produce propionate from a range of substrates and via different pathways, e. g. Propionibacterium sp. via the Wood-Werkman cycle, or Clostridium and Megasphaera sp. via the acrylate pathway (Gonzalez-Garcia et al. 2017). Especially Propionibacterium sp. such as Propionibacterium acidipropionici and Propionibacterium freudenreichii have been studied extensively to develop fermentation strategies that allow the turnover of cheap substrates such as glycerol or molasses to propionate (Dishisha et al. 2012; Feng et al. 2011). Furthermore, these bacteria have also been engineered to improve propionate yields and overcome typical fermentation obstacles such as low acid tolerance (Jiang et al. 2015; Wang et al. 2015). Aside from propionibacteria, other non-native propionate-producing microorganisms have been engineered for propionate production, including Escherichia coli (Akawi et al. 2015; Gonzalez-Garcia et al. 2020; Kandasamy et al. 2013), Lactobacillus plantarum (Balasubramanian and Subramanian 2019), and Pseudomonas putida (Ma et al. 2020; Mu et al. 2021). Surprisingly, clostridia have never been considered hosts for recombinant propionate production although they are organisms with a versatile metabolism enabling them to use diverse carbon sources, including lignocellulosic hydrolysates and waste-derived substrates, and convert them into various products (Cho et al. 2015; Tracy et al. 2012). Furthermore, multiple tools are available to genetically modify clostridia for optimized production of native or recombinant compounds thus making them promising host strains for the production of commodity chemicals such as ethanol, isopropanol, 2,3-butanediol, or fatty acid esters (Cho et al. 2015; Feng et al. 2021). Clostridium saccharoperbutylacetonicum is a well-characterized solventogenic bacterium, which is genetically accessible and has high growth rates in favorable medium. Since it is a known hyper-butanol producer, it has mostly been employed for butanol production (Jiménez-Bonilla et al. 2021). However, C. saccharoperbutylacetonicum has also successfully been used for the production of hydrogen (Singh et al. 2019), isopropanol (Wang et al. 2019), 1,3-butanediol (Grosse-Honebrink et al. 2021), as well as caproate and hexanol (Wirth and Dürre 2021), thus highlighting its potential as a host for production of recombinant compounds. Here, we report the approach to convert C. saccharoperbutylacetonicum into a propionate producer by the implementation of the acrylate pathway from An. neopropionicum and a D-lactate dehydrogenase from L. mesenteroides subsp. mesenteroides (Fig. 1). For that purpose, a two-plasmid system harboring two propionate-synthesis operons (PSOs) was constructed, and gene expression was controlled by the sigma factor-inducible PtcdB promoter from C. difficile. In order to identify promoters that are suitable for mediation of gene expression in C. saccharoperbutylacetonicum, a promoter study using FAST was conducted.
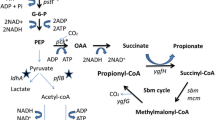
Schematic overview of glycolytic and acidogenic pathways in C. saccharoperbutylacetonicum based on Jones and Woods (1986) coupled with acrylate pathway for propionate production (grey box; based on Hetzel et al. 2003) from An. neopropionicum and D-lactate dehydrogenase from L. mesenteroides subsp. mesenteroides (stoichiometrically incorrect). LdhD, D-lactate dehydrogenase (LEUM_1756); Pct, propionate CoA-transferase (CLNEO_17700); Lcd, lactoyl-CoA dehydratase (CLNEO_17730-17710); Acr, acryloyl-CoA reductase (CLNEO_21740-21760); ABE, acetone-butanol-ethanol
Materials and methods
Bacterial strains and cultivation
Bacterial strains and plasmids used are listed in Table 1. E. coli XL1-Blue MRF´ was used for cloning purposes and cultivated aerobically under constant shaking (180 rpm) at 37 °C in Luria–Bertani (LB) medium (Green and Sambrook 2012) supplemented with respective antibiotics. For the preparation of chemically competent cells, E. coli was grown in Super Optimal Broth (SOB; Hanahan 1983) and cultivated at 18 °C with shaking (100 rpm). C. saccharoperbutylacetonicum N1-4(HMT) DSM 14923 was used as a production host for propionate and cultivated under strictly anaerobic conditions at 30 °C in complex (clostridial growth medium, CGM) or minimal medium (optimized synthetic medium, OMS). Both media were prepared as previously described by Wirth and Dürre (2021); however, sodium sulfide was omitted from OMS. When C. saccharoperbutylacetonicum was cultivated on solid medium, CGM was supplemented with 1.5% agar (w v−1) and antibiotics. The medium was prepared and poured within an anaerobic cabinet with a N2:H2 (95:5%) gas atmosphere using anaerobic water. Cells of C. saccharoperbutylacetonicum grown on solid CGM were cultivated in an incubator located in the anaerobic cabinet at 32 °C. Antibiotics used to select recombinant bacterial strains were supplemented to the following concentrations (per milliliter): 30 µg chloramphenicol, 10 µg clarithromycin, 300 µg erythromycin, 10 µg tetracycline, 40 µg thiamphenicol.
Growth experiments for propionate production as well as fluorescence determination were performed in 50 ml OMS in 125-ml Müller-Krempel bottles (Müller & Krempel AG, Bülach, Switzerland) supplemented with respective antibiotics. In the case of propionate production, 2-ml samples were withdrawn throughout the course of growth for the determination of substrate consumption and product formation. For determination of promoter activities, 0.5–2-ml samples were withdrawn at different stages of growth and processed as described in the “FAST reporter assays” section.
Plasmid construction
Standard molecular cloning techniques were performed according to established protocols (Green and Sambrook 2012). Linearization of plasmids was performed using “FastDigest™ enzymes” (Thermo Fisher Scientific Inc., Waltham, MA, USA). Primers used for amplification of genes and promoters are listed in Table 2 and were synthesized by biomers.net GmbH (Ulm, Germany). Amplification of DNA fragments was performed using the “CloneAmp™ HiFi polymerase” (Takara Bio USA, Inc., Mountain View, CA, USA) or the “Phusion™ Green High-Fidelity DNA polymerase” (Thermo Fisher Scientific Inc., Waltham, MA, USA). Generated DNA fragments were purified using the “Zymoclean™ Gel DNA Recovery Kit” (ZYMO Research Corp., Irvine, CA, USA) according to the manufacturer´s instructions. All plasmids were constructed using either the “In-Fusion® HD Cloning kit” (Takara Bio USA, Inc., Mountain View, CA, USA) or the “NEBuilder® HiFi DNA Assembly Master Mix” (New England Biolabs® Inc., Ipswich, MA, USA) following manufacturer´s instructions. A total of 8–10 µl of cloning mixtures were finally used to transform chemically competent E. coli cells. Plasmid DNA from E. coli strains was isolated using the “Zyppy™ Plasmid Miniprep Kit” (ZYMO Research Corp., Irvine, CA, USA) following the manufacturer´s instructions.
To establish propionate production in C. saccharoperbutylacetonicum, respective genes encoding the key enzymes of the acrylate pathway from An. neopropionicum were used (Beck et al. 2016). These include the propionate CoA-transferase (pct; CLNEO_17700), the subunits of the lactoyl-CoA dehydratase (lcdC, lcdA, lcdB; CLNEO_17730-17710), and the subunits of the acryloyl-CoA reductase (acrC, acrB, acrA; CLNEO_21740-21760). Furthermore, the well-characterized D-lactate dehydrogenase gene from L. mesenteroides subsp. mesenteroides (ldhD; LEUM_1756; Li et al. 2012) was inserted into the PSO. Previously, all mentioned genes except for acrA, acrB, and acrC were assembled on plasmid pMTL83151_Ptet_3-HP_opt (Beck 2020) and controlled by the tetracycline-inducible promoter Ptet (comprises Pxyl from Bacillus subtilis as well as tetR and PtetR from Tn10 from E. coli; Zhang et al. 2000; Fagan and Fairweather 2011; Beck et al. 2020). The aforementioned genes including Ptet were PCR-amplified from pMTL83151_Ptet_3-HP_opt using primers Inf_tetR-pct_fw and Inf_tetR-pct_rev and subcloned in pMTL83151, which was linearized with XhoI and SgsI (resulting in pMTL83151_Ptet_LLP). In a second step, genes acrA, acrB, and acrC were amplified from genomic DNA of An. neopropionicum DSM 3847 using primers Inf_acr_fw2 and Inf_acr_rev2 and inserted into pMTL83151_Ptet_LLP digested with XhoI and NotI to yield pMTL83151_Ptet_LLPA. This plasmid could successfully be constructed; however, DNA concentration was low, and when the respective E. coli strain was inoculated to harvest enough DNA for transformation of C. saccharoperbutylacetonicum, the plasmid was altered. Therefore, genes encoding the acrylate pathway were divided into two PSOs and assembled on two plasmids, which were based on vectors pMTL83151 and pMTL82251 (Heap et al. 2009). PSO1 consisted of genes pct, acrC, acrB, and acrA (pMTL83151_PA), PSO2 harbored ldhD, lcdC, lcdA, and lcdB (pMTL82251_LL). For cloning of pMTL83151_PA and pMTL82251_LL, plasmids pMTL83151 and pMTL82251 were digested using enzymes XhoI and NheI and ligated with PCR-amplified fragments pct-acrCBA (from pMTL83151_Ptet_LLPA; primers Inf_pct-acrCBA_fw and Inf_pct-acrCBA_rev) and ldhD-lcdCAB (from pMTL83151_Ptet_LLP; primers Inf_ldhD-lcdCAB_fw and Inf_ldhD-lcdCAB_rev), respectively. To control the expression of both PSOs, the PtcdB-tcdR promoter system from C. difficile was selected since this promoter system should be tight in E. coli (Moncrief et al. 1997). Therefore, PtcdB was inserted upstream of pct-acrCBA on pMTL83151_PA. Plasmid pMTL83151_PA was digested using Eco72I and SdaI and ligated with PtcdB amplified from genomic DNA from C. difficile DSM 27147 using primers Inf_PtcdB_fw and Inf_PtcdB_rev to construct pMTL83151_PtcdB_PA. The alternative sigma factor needed for PtcdB recognition (TcdR; Martin-Verstraete et al. 2016) is provided on plasmid pMTL82251_LL and was added to the existing operon on this plasmid together with the lactose-inducible promoter PbgaL from C. perfringens (Hartman et al. 2011) to control the expression of the PSO finally consisting of ldhD, lcdC, lcdA, lcdB, and tcdR (pMTL82251_PbgaL_LL_tcdR). To construct pMTL82251_PbgaL_LL_tcdR, plasmid pMTL82251_LL was linearized using XhoI and SdaI and fused with Ptet amplified from pMTL83151_Ptet_3-HP_opt using primers Inf_Ptet_fw and Inf_Ptet_LL_rev. Resulting plasmid pMTL82251_Ptet_LL was cut using NheI, and the generated backbone was used in a cloning reaction together with amplified tcdR (primers Inf_tcdR_wRBS_fw and Inf_tcdR_wRBS_rev; template genomic DNA from C. difficile DSM 27147) to build pMTL82251_Ptet_LL_tcdR. For construction of pMTL82251_PbgaL_LL_tcdR, PbgaL was amplified from plasmid pMTL83151_gusA_PbgaL (Beck et al. 2020) using primers Inf_PbgaL_fw and Inf_PbgaL_rev and inserted in linearized pMTL82251_Ptet_LL_tcdR (linearized using enzymes XhoI and SalI) to exchange Ptet against PbgaL. The resulting two-plasmid system is displayed in Figure S1.
Existing PSOs were reconstructed to even out the proposed imbalance in the acrylate pathway for optimization of propionate concentrations. Therefore, pct was moved to PSO2 (controlled by PbgaL instead of PtcdB) whereas genes lcdC, lcdA, and lcdB were moved to the space formerly taken by pct in PSO1 (controlled by PtcdB instead of PbgaL). Also, a second PtcdB and a second acr gene cluster were inserted in PSO1 in two separate cloning steps (plasmids pMTL83151_PtcdB_L_PtcdB and pMTL83151_PtcdB_L_PtcdB_AA, respectively). Additionally, PSO2 was extended by another tcdR gene to yield pMTL82251_PbgaL_LPTT. To construct the aforementioned plasmids, pMTL83151_PtcdB_PA was cut using XbaI and NheI, and pMTL82251_PbgaL_LL_tcdR was digested using XmaJI and NheI. The genes and promoters to be relocated or inserted a second time were amplified using primers Inf_pct_fw and Inf_pct_rev (pct; template pMTL83151_PtcdB_PA), Inf_lcd_fw and Inf_lcd_rev (lcdCAB; template pMTL82251_LL), Inf_tcdR_fw2 and Inf_tcdR_rev2 (tcdR; template pMTL82251_PbgaL_LL_tcdR), as well as Inf_PtcdB_fw2 and Inf_PtcdB_rev2 (PtcdB; template pMTL83151_PtcdB_PA) and fused with generated backbones to build plasmids pMTL83151_PtcdB_L_PtcdB and pMTL82251_PbgaL_LPTT. In a second step, pMTL83151_PtcdB_L_PtcdB was linearized using enzymes XhoI and SgsI and ligated with two acr gene clusters amplified from pMTL83151_PtcdB_PA using primers Inf_acr_fw3 and Inf_acr_rev3 (acr cluster 1) as well as Inf_acr_fw4 and Inf_acr_rev4 (acr cluster 2) to construct pMTL83151_PtcdB_L_PtcdB_AA. The resulting plasmids are shown in Figure S2.
All plasmids constructed for the evaluation of promoter activities in C. saccharoperbutylacetonicum were based on the pMTL83251 backbone (Heap et al. 2009). Plasmid pMTL83251_Ppta-ack_FAST was kindly provided by Maximilian Flaiz (University of Ulm, unpublished). To construct further plasmids harboring feg (FAST-encoding gene) as a reporter gene, pMTL83251_PbgaL_FAST (Flaiz et al. 2021) was linearized using BamHI and NdeI and fused with PCR-amplified promoters Pbld (from C. saccharoperbutylacetonicum), PtcdB (from C. difficile), and PlctB (from Acetobacterium woodii). Templates for amplification of promoters were genomic DNA from C. saccharoperbutylacetonicum N1-4(HMT) DSM 14923 (Pbld; primers Inf_Pbld_FAST_fw and Inf_Pbld_FAST_rev), plasmid pMTL83151_PtcdB_PA (PtcdB; primers Inf_PtcdB_FAST_fw and Inf_PtcdB_FAST_rev), and genomic DNA from A. woodii DSM 1030 (PlctB; primers Inf_PlctB_FAST_fw and Inf_PlctB_FAST_rev). Final plasmids were pMTL83251_Pbld_FAST, pMTL83251_PtcdB_FAST, and pMTL83251_PlctB_FAST, respectively. To monitor the activity of PtcdB, plasmids carrying feg under control of PtcdB and tcdR controlled by either Pbld or PbgaL were constructed. For that purpose, pMTL83251_PtcdB_FAST was digested using NotI and NdeI and ligated with fragments PbgaL and tcdR (both amplified from pMTL82251_PbgaL_LL_tcdR using primers Inf_PbgaL_tcdR-fdx_fw and Inf_PbgaL_tcdR-fdx_rev (PbgaL) as well as Inf_tcdR-fdx_fw and Inf_tcdR-fdx_rev (tcdR)) to yield pMTL83251_PtcdB_FAST_PbgaL_tcdR. Then, pMTL83251_PtcdB_FAST_PbgaL_tcdR was linearized with SmaI and SalI and PbgaL was exchanged against Pbld (amplified from pMTL83251_Pbld_FAST with primers Inf_Pbld_fw2 and Inf_Pbld_rev2) to assemble plasmid pMTL83251_PtcdB_FAST_Pbld_tcdR.
Transformation of bacterial strains
Preparation and transformation of chemically competent E. coli cells were performed following procedures previously described by Weitz et al. (2021). Transformation of C. saccharoperbutylacetonicum was performed using electroporation. Electrocompetent cells were prepared following known procedures (Wirth and Dürre 2021) with slight modifications. Cells were cultivated at 30 °C until mid-exponential growth phase (OD600 of 0.8–1.2). Centrifugation steps were performed in an anaerobic cabinet for 10 min at 3,985 × g and 4 °C. Recovery of pulsed cells was performed for 2–16 h at 32 °C.
Analytical methods
The growth of bacterial strains was monitored by measuring the optical density at a wavelength of 600 nm (OD600) using the “Ultrospec™ 3100 pro UV/Visible” spectrophotometer (Amersham Biosciences Europe GmbH, Freiburg, Germany).
Quantification of metabolic products acetone, ethanol, propanol, butanol, acetate, propionate, and butyrate was achieved using gas chromatography (GC). In total, 2-ml samples were withdrawn from cultures throughout the course of growth experiments and centrifuged (15,000 × g, 30 min, 4 °C). A total of 480 µl of supernatant were filled into 2-ml crimp vials (CS-Chromatographie Service GmbH, Langerwehe, Germany), acidified by addition of 20 µl 2 M HCl, and closed with aluminum caps. Prepared samples were analyzed using a “Clarus 600” gas chromatograph (PerkinElmer, Inc., Waltham, MA, USA) equipped with an “Elite-FFAP” capillary column (inner diameter 0.32 mm × 30 m) and a flame ionization detector operating at 300 °C. H2 served as carrier gas with a flow rate of 2.25 ml min−1, injection temperature was set to 225 °C, detector gases were H2 (45 ml min−1) and synthetic air (79.5% N2 + 20.5% O2 at 450 ml min−1). 1 µl of culture supernatant was injected into the gas chromatograph and analyzed using the following temperature profile: 80 °C for 2 min followed by a gradual increase of temperature to 190 °C with a rate of 10 °C min−1, then, the temperature was increased to 250 °C at 40 °C min−1, and finally 250 °C were held constant for 1 min. For calibration purposes, defined external standards containing all substances were prepared.
Glucose consumption and lactate production were determined via high-performance liquid chromatography (HPLC) using the “Infinity 1260” HPLC system (Agilent Technologies, Santa Clara, CA, USA) equipped with a “CS-Organic-Acid Resin column” (150 × 8 mm; CS-Chromatographie Service GmbH, Langerwehe, Germany), a refraction index detector (for glucose) operating at 35 °C, and a diode array UV detector (for lactate) operating at a wavelength of 210 nm and room temperature. Culture supernatants were prepared as described for GC; however, no acidification of samples was necessary. As mobile phase, 5 mM H2SO4 with a flow rate of 0.7 ml min−1 was used. The sample volume injected into the HPLC system was 20 µl, and external standards again were prepared for calibration of determined compounds.
FAST reporter assays
To monitor fluorescence levels of FAST-producing strains, 0.5–2-ml samples were withdrawn from cultures at different stages of growth and processed as described by Flaiz et al. (2021) with one exception. Washing steps were carried out for 10 min at 7,607 × g and 4 °C. To determine the fluorescence intensities (FLU) of FAST-producing cultures, a microplate reader was used. Fluorescence was determined following the established protocol of Flaiz et al. (2021) with one exception. Instead of 10 µM TFLime, only 5 µM TFLime were supplemented to PBS-suspended cells. FLU of cells without the addition of TFLime was determined as a negative control. Finally, fluorescence intensities were normalized to OD600 of PBS-suspended cells. Normalized FLU without the addition of TFLime were subtracted from FLU with TFLime to determine the actual FLU of the culture. Monitoring of FLU at single-cell level was performed via flow cytometry following procedures as described by Flaiz et al. (2022). Staining of washed cells was achieved by the addition of 5 µM TFLime (final OD600: 0.08).
Results
Construction of a two-plasmid system for propionate production
The construction of a PSO harboring all acrylate pathway genes and ldhD was not possible due to recurring mutations detected in the acrC gene upon inoculation of the respective E. coli strain from conserved stocks. Various cloning strategies using different promoters, origins of replication, and E. coli cloning hosts failed (data not shown). Therefore, the PtcdB-tcdR promoter system from C. difficile was chosen to tightly control the expression of the acr gene cluster. The PtcdB promoter is dependent on induction with an alternative sigma factor (TcdR) as its -10 region does not contain the typical TATA motif and therefore cannot be recognized by standard bacterial sigma factors. Only when TcdR interacts with the RNA polymerase, transcription from PtcdB can be initiated (Mani and Dupuy 2001; Martin-Verstraete et al. 2016). Thus, as long as TcdR is not provided in the same cell as PtcdB, expression of the genes under the control of PtcdB should not be possible. To make use of this unique promoter system, the originally planned PSO was divided into two parts and assembled on a two-plasmid system (Fig. S1). PSO1 consisting of the acr gene cluster as well as the pct gene was put under control of PtcdB, whereas PSO2, which harbored genes ldhD, lcdA, lcdB, and lcdC, was extended by the tcdR gene. This way, leaky expression of acrC in E. coli could be prevented as no mutations were detected and cloning of both plasmids was successful (data not shown).
Determination of promoter activities using FAST
Since PtcdB needs to be induced by the alternative sigma factor TcdR, the expression of tcdR has to be controlled separately. A promoter study using FAST as a fluorescent reporter was conducted to assess the strength and activity pattern of different clostridial promoters throughout the growth of C. saccharoperbutylacetonicum. This way, suitable promoters to drive tcdR expression should be identified. An overview of tested promoters as well as their known characteristics is given in Table 3. Figure 2 summarizes the maximal fluorescence intensities (FLU) detected for all promoters screened normalized to the OD600. The highest activity was measured for the promoter Pbld from C. saccharoperbutylacetonicum as the respective strain reached a FLU OD600−1 of 60,000. With this value, FLU of C. saccharoperbutylacetonicum [pMTL83251_Pbld_FAST] was more than twice as high as the FLU of C. saccharoperbutylacetonicum expressing feg under control of PthlA from C. acetobutylicum (27,600) and approx. six-fold higher than FLU of the induced C. saccharoperbutylacetonicum [pMTL83251_PbgaL_FAST] (9,500). The strain C. saccharoperbutylacetonicum [pMTL83251_Ppta-ack_FAST], which carried the Ppta-ack promoter from C. ljungdahlii, was only weakly fluorescent. Its maximal FLU value of 3,100 was the lowest FLU measured for any strain aside from the autofluorescence displayed by C. saccharoperbutylacetonicum strains without feg expression, i.e., C. saccharoperbutylacetonicum [pMTL83251] and non-induced C. saccharoperbutylacetonicum strains with feg under control of PbgaL from C. perfringens or PlctB from A. woodii, respectively. The strain C. saccharoperbutylacetonicum [pMTL83251_PlctB_FAST] also only showed autofluorescence when induced with D- or L-lactate indicating that PlctB is not active in C. saccharoperbutylacetonicum. Expression profiles of the strains revealed that all promoters have dynamic activities as FLU of all strains first rose during exponential growth phases and subsided with ongoing cultivation (Fig. S3). Only Ppta-ack showed a steady activity as FLU was constant between the mid-exponential and the mid-stationary phase. Furthermore, the two strongest promoters of this study, Pbld and PthlA, were confirmed to have an exponential growth phase-associated activity.
Maximal fluorescence intensities measured for FAST-producing C. saccharoperbutylacetonicum strains. All strains carried pMTL83251-based plasmids with feg under control of different promoters. Control, pMTL83251 without feg; Ppta-ack, feg under control of Ppta-ack from C. ljungdahlii; PthlA, feg under control of PthlA from C. acetobutylicum; Pbld, feg under control of Pbld from C. saccharoperbutylacetonicum; PbgaL, feg under control of PbgaL from C. perfringens (n. i., non-induced; i., induced using 20 mM lactose); PlctB, feg under control of PlctB from A. woodii (n. i., non-induced; i., induced using 15 mM D- or L-lactate, respectively). Fluorescence intensities (FLU) were normalized to OD600 and are given in arbitrary units (a. u.). Error bars indicate standard deviations, n = 3
In a second step, the activity of PtcdB was investigated in dependence on different levels of tcdR expression. For that purpose, recombinant C. saccharoperbutylacetonicum strains carrying plasmids with feg under the control of PtcdB and tcdR under the control of either Pbld or PbgaL were constructed. Promoters Pbld and PbgaL were chosen due to the bright fluorescence of respective strains. Also, PbgaL tightly regulated gene expression as FLU was only detected in the induced culture (Fig. 2). Subsequently, PtcdB activity was investigated at both whole-culture and single-cell level using a microplate reader and flow cytometry. As negative controls, strains harboring the empty vector pMTL83251 or a plasmid with feg under control of PtcdB but without a tcdR gene (pMTL83251_PtcdB_FAST) were cultivated in parallel to strains C. saccharoperbutylacetonicum [pMTL83251_PtcdB_FAST_Pbld_tcdR] and C. saccharoperbutylacetonicum [pMTL83251_PtcdB_FAST_PbgaL_tcdR]. As previously observed, FLU of FAST-producing strains was highly dynamic during growth of C. saccharoperbutylacetonicum (Fig. 3a). While the negative controls only showed autofluorescence, FLU of C. saccharoperbutylacetonicum [pMTL83251_PtcdB_FAST_Pbld_tcdR] and induced C. saccharoperbutylacetonicum [pMTL83251_PtcdB_FAST_PbgaL_tcdR] increased during exponential growth phases and reached a maximum during early stationary phases. The maximum FLU of the strain with tcdR controlled by Pbld was approx. 28-fold higher than the autofluorescence of the negative controls and three-fold higher than FLU of the strain with lactose-inducible tcdR expression. With ongoing cultivation, FLU of both strains declined to levels comparable to empty vector and non-induced strains. Flow cytometric analyses of the FAST-producing strains revealed heterogenous populations of fluorescent and non-fluorescent C. saccharoperbutylacetonicum cells (Fig. 3b). While C. saccharoperbutylacetonicum [pMTL83251_PtcdB_FAST] did not show any fluorescence besides the previously described autofluorescence, induction of tcdR expression with lactose resulted in a shift of the population, indicating fluorescence due to an activated PtcdB. Similarly, cells with autonomous tcdR expression (Pbld control), and thus, autonomic PtcdB induction became fluorescent as the population shifted in direction of increasing FLU. After 64 h of cultivation, the maximal amounts of fluorescent cells were detected for both FAST-producing cultures, which accounted for 29% and 42% for PbgaL- and Pbld-controlled tcdR expression, respectively. Additional data on flow cytometric analyses of the strains including the investigation of FAST production in strains C. saccharoperbutylacetonicum [pMTL83251_Pbld_FAST] and C. saccharoperbutylacetonicum [pMTL83251_PbgaL_FAST] are shown in Figure S4. When feg expression was directly controlled by Pbld or PbgaL, the maximum amounts of fluorescent cells were significantly higher compared to feg expression controlled by PtcdB resulting in a total of 97% and 76% of fluorescent cells, respectively. In later stages of growth, FLU of all strains decreased as populations became non-fluorescent. Although PtcdB activity was higher when tcdR expression was under control of Pbld, the lactose-inducible PbgaL promoter was chosen to establish propionate production in C. saccharoperbutylacetonicum to avoid possible pathway overloading due to the high activity observed for Pbld (Grosse-Honebrink et al. 2021). Furthermore, the use of PbgaL allows a controlled induction of gene expression and has recently successfully been used to establish a two-plasmid system carrying PtcdB and tcdR in C. saccharoperbutylacetonicum (Flaiz et al. 2022).
Evaluation of PtcdB activity in C. saccharoperbutylacetonicum using FAST assay. All strains carried pMTL83251-based plasmids with feg under control of PtcdB from C. difficile and tcdR under control of PbgaL from C. perfringens or Pbld from C. saccharoperbutylacetonicum. a Growth (solid lines) and fluorescence intensities (dashed lines) of C. saccharoperbutylacetonicum [pMTL83251] (circles), C. saccharoperbutylacetonicum [pMTL83251_PtcdB_FAST] (right-facing triangles), C. saccharoperbutylacetonicum [pMTL83251_PtcdB_FAST_PbgaL_tcdR] with (crossed squares) and without (half-filled squares) induction of gene expression, and C. saccharoperbutylacetonicum [pMTL83251_PtcdB_FAST_Pbld_tcdR] (half-filled downward-facing triangles). b Density plots of C. saccharoperbutylacetonicum [pMTL83251_PtcdB_FAST] (left), induced C. saccharoperbutylacetonicum [pMTL83251_PtcdB_FAST_PbgaL_tcdR] (middle), and C. saccharoperbutylacetonicum [pMTL83251_PtcdB_FAST_Pbld_tcdR] (right) after 64 h of cultivation. Black dashed line indicates time of induction of PbgaL using 20 mM lactose. Error bars indicate standard deviations, n = 3
Establishment of propionate production in C. saccharoperbutylacetonicum
As previously mentioned, a two-plasmid system carrying the two PSOs was constructed to convert C. saccharoperbutylacetonicum into a propionate producer (Fig. S1). These two plasmids were used to transform C. saccharoperbutylacetonicum, resulting in the strain C. saccharoperbutylacetonicum [pMTL83151_PtcdB_PA][pMTL82251_PbgaL_LL_tcdR]. As a negative control, a strain carrying the empty vectors pMTL83151 and pMTL82251 was constructed. Recombinant strains were then tested in comparison to C. saccharoperbutylacetonicum wild type in minimal medium using glucose as a carbon source (Fig. 4). The strain harboring the PSOs showed considerable differences in growth and production behavior compared to the control strains. Aside from a strongly diminished growth and incomplete glucose consumption, acetate was accumulated in both induced and non-induced C. saccharoperbutylacetonicum [pMTL83151_PtcdB_PA][pMTL82251_PbgaL_LL_tcdR] cultures (30 mM and 42.1 mM). Butyrate formation of C. saccharoperbutylacetonicum strains carrying PSOs was comparable to the control strains; however, reassimilation of this acid was delayed by 44 h. The induced culture of C. saccharoperbutylacetonicum [pMTL83151_PtcdB_PA][pMTL82251_PbgaL_LL_tcdR] produced 27.5 mM lactate, 0.7 mM propionate, and 0.8 mM propanol, whereas the controls and the non-induced strain only produced little lactate (4.7 to 6.9 mM) and no propionate or propanol. This provided evidence for the first heterologous production of propionate achieved in any Clostridium sp. Solvent production of C. saccharoperbutylacetonicum strains carrying the PSOs was delayed and reduced compared to the control strains, which is also reflected in the pH development. While the pH of C. saccharoperbutylacetonicum wild type and the vector control strain increased to values of 6 and 5.9 after a drop during the first 37 h of cultivation, pH of C. saccharoperbutylacetonicum [pMTL83151_PtcdB_PA][pMTL82251_PbgaL_LL_tcdR] dropped to a value of 5.6 and only slightly increased after 97 h. Final acetone titers of non-induced and induced C. saccharoperbutylacetonicum [pMTL83151_PtcdB_PA][pMTL82251_PbgaL_LL_tcdR] were in the range of the vector control and wild type strains, respectively. However, only half the amount of ethanol and butanol were produced by the PSO-carrying strains.
Results of growth experiment to establish propionate production in C. saccharoperbutylacetonicum. Growth (OD600), pH, glucose consumption, and product formation were monitored throughout the course of the experiment. Open circles, C. saccharoperbutylacetonicum wild type; squares, C. saccharoperbutylacetonicum [pMTL83151][pMTL82251]; open triangles, non-induced C. saccharoperbutylacetonicum [pMTL83151_PtcdB_PA][pMTL82251_PbgaL_LL_tcdR]; filled triangles, induced C. saccharoperbutylacetonicum [pMTL83151_PtcdB_PA][pMTL82251_PbgaL_LL_tcdR]. Induction of gene expression was achieved by addition of 20 mM lactose and is indicated with a black dashed line. Dotted lines indicate ethanol or propanol production of respective strains. Error bars represent standard deviations, n = 3
Optimization of propionate production in C. saccharoperbutylacetonicum
The productivity of the recombinant strain C. saccharoperbutylacetonicum [pMTL83151_PtcdB_PA][pMTL82251_PbgaL_LL_tcdR] with an induced gene expression was rather low as only 0.7 mM propionate was detected (Fig. 4). Since the two PSOs were under the control of two promoters with different strengths (PtcdB weaker than PbgaL), a metabolic imbalance was proposed as the reason for the low productivity. To even out this postulated imbalance and ensure a more balanced carbon flux through the recombinant pathway, the two PSOs were reconstructed. The resulting new plasmid system carried two copies of the acr and one copy of the lcd gene cluster under the control of separate PtcdB, as well as the pct, the ldhD, and two tcdR genes under the control of PbgaL (Fig. S2). The new PSOs were used to transform C. saccharoperbutylacetonicum to build the strain C. saccharoperbutylacetonicum [pMTL83151_PtcdB_L_PtcdB_AA][pMTL82251_PbgaL_LPTT]. This strain was then characterized in a growth experiment in comparison to the original producer C. saccharoperbutylacetonicum [pMTL83151_PtcdB_PA][pMTL82251_PbgaL_LL_tcdR] as well as the negative controls C. saccharoperbutylacetonicum [pMTL83151][pMTL82251] and C. saccharoperbutylacetonicum wild type (Fig. 5). Recombinant strains carrying old and new PSOs showed impaired growth and did not deplete the provided glucose. The induced strain carrying the optimized PSOs produced up to 26.1 mM acetate, 42.4 mM butyrate, 57.9 mM lactate, and 3 mM propionate. Solvent production was limited to traces of acetone (0.2 mM) and propanol (0.1 mM) as well as small amounts of ethanol (1.3 mM) and butanol (6.8 mM). Aside from a lactate uptake, no acid reassimilation was observed for this strain. The non-induced C. saccharoperbutylacetonicum [pMTL83151_PtcdB_L_PtcdB_AA][pMTL82251_PbgaL_LPTT] also accumulated acids rather than solvents; however, this strain produced less lactate and more acetate and butyrate. C. saccharoperbutylacetonicum harboring the original PSOs showed a growth and production profile similar to the growth experiment displayed in Fig. 4. Again, acetate was accumulated, and butyrate was reassimilated with a delay compared to the control strains. Lactate production of the induced C. saccharoperbutylacetonicum [pMTL83151_PtcdB_PA][pMTL82251_PbgaL_LL_tcdR] strain was elevated (25.2 mM), propionate concentration reached a maximum of 0.8 mM. Moreover, 0.5 mM propanol was detected. Further solvents produced by non-induced and induced C. saccharoperbutylacetonicum [pMTL83151_PtcdB_PA][pMTL82251_PbgaL_LL_tcdR] were acetone (up to 43.3 mM), ethanol (up to 8.1 mM), and butanol (up to 84.5 mM). A direct comparison of the original and the optimized C. saccharoperbutylacetonicum strains revealed an increase in propionate production of 375%. Thus, the reconstruction of the PSOs led to an almost four-fold increase in propionate.
Results of growth experiment to improve propionate production in C. saccharoperbutylacetonicum. Growth (OD600), pH, glucose consumption, and product formation were monitored throughout the course of the experiment. Open circles, C. saccharoperbutylacetonicum wild type; squares, C. saccharoperbutylacetonicum [pMTL83151][pMTL82251]; open triangles, non-induced C. saccharoperbutylacetonicum [pMTL83151_PtcdB_PA][pMTL82251_PbgaL_LL_tcdR]; filled triangles, induced C. saccharoperbutylacetonicum [pMTL83151_PtcdB_PA][pMTL82251_PbgaL_LL_tcdR]; open diamonds, non-induced C. saccharoperbutylacetonicum [pMTL83151_PtcdB_L_PtcdB_AA][pMTL82251_PbgaL_LPTT]; filled diamonds, induced C. saccharoperbutylacetonicum [pMTL83151_PtcdB_L_PtcdB_AA][pMTL82251_PbgaL_LPTT]. Induction of gene expression was achieved by addition of 20 mM lactose and is indicated with dashed lines. Dotted lines indicate ethanol or propanol production of respective strains. Error bars represent standard deviations, n = 3
Discussion
The data presented clearly show that heterologous expression of the acrylate pathway leads to propionate production in C. saccharoperbutylacetonicum. In the first growth experiment, only traces of propionate (0.7 mM) were obtained, indicating that the carbon flux through the established pathway was not high. Although induction of gene expression led to elevated lactate concentrations in comparison to control strains, possibly a result of the heterologously expressed D-lactate dehydrogenase, lactate turnover to propionate seemed to be limited. The acrylate pathway consists of multiple steps starting with the activation of D-lactate to its CoA-derivative D-lactoyl-CoA, which is subsequently dehydrated to the toxic compound acryloyl-CoA. These reactions are catalyzed by the propionate CoA-transferase and lactoyl-CoA dehydratase, respectively. The resulting acryloyl-CoA is reduced to propionyl-CoA by an acryloyl-CoA reductase. Finally, propionate is released as the product by a CoA transfer from propionyl-CoA to D-lactate by the Pct (Fig. 1; Hetzel et al. 2003). Considering the fact that the PSOs encoding all mentioned enzymes were controlled by two different promoters, which FAST studies showed to be highly different in strength (Figs. 2, 3, and S4), the concern of a metabolic imbalance is valid. Lcd-encoding genes were under the control of the stronger PbgaL promoter, thus possibly leading to a higher expression level compared to the acr genes, which were controlled by the weaker PtcdB promoter. Based on these assumptions it would follow that the Lcd turnover is higher than the Acr turnover, thereby leading to an acryloyl-CoA accumulation. Furthermore, the Acr apparently has a low catalytic efficiency, which, according to reports is so low that native producers must compensate for this by producing high amounts of the enzyme (Hetzel et al. 2003; Kandasamy et al. 2013). Therefore, it is likely that acryloyl-CoA was accumulated rather than converted by Lcd and Acr reactions. Due to the electrophilic properties of acryloyl-CoA (Herrmann et al. 2005), it seems logical that an accumulation of this toxic compound would cause a high stress level for the production host, which could manifest itself in an impaired growth, a delayed or complete lack of acid reassimilation and solventogenesis, and a low productivity, all of which was observed in the growth experiments conducted in this study. Also, since the acrylate pathway is an electron sink, a deficiency in reducing equivalents could be a consequence (Kandasamy et al. 2013). With the metabolism out of balance due to the high burden exerted by the accumulation of pathway intermediates such as acryloyl-CoA and depletion of the NADH pool, high strain performances cannot be expected. Furthermore, FAST studies conducted via flow cytometry revealed that C. saccharoperbutylacetonicum cultures were heterogeneous even in the case of feg expression mediated by the strong and native Pbld promoter (Figs. 3 and S4). Especially the use of the PtcdB-tcdR system with tcdR expression driven by PbgaL, which reflects the situation created in the propionate-producing strains, seems to limit the amount of productive cells as only 29% showed fluorescence in presence of TFLime (Fig. 3b). When transferring this observation to the strains carrying PSOs under control of PtcdB, it is reasonable to assume that only a small portion of the culture expressed the acrylate pathway genes and thus was able to form propionate. Such heterogeneity of bacterial populations in connection with an induced, plasmid-based gene expression has been shown several times and hypothesized to be caused by the uptake mechanism and concentration of the inducer, plasmid instability, or plasmid loss (Binder et al. 2016; Flaiz et al. 2021; Siegele and Hu 1997). Further factors to contribute to culture heterogeneity are cell morphogenesis or sporulation, both of which are especially apparent in clostridial cultures (Jones et al. 2008; Tracy et al. 2010). Most recently, Flaiz et al. (2022) also demonstrated heterogeneity of C. saccharoperbutylacetonicum cultures expressing feg in a PtcdB-dependent manner with tcdR controlled by PbgaL. Again, culture heterogeneity was observed throughout all growth phases, although in this case, the number of fluorescent cells was higher compared to the present study (Flaiz et al. 2022). Nevertheless, the impact of culture heterogeneity on the production behavior of C. saccharoperbutylacetonicum cannot be neglected and has to be considered a major contributor to the low observed productivity aside from the postulated metabolic imbalance.
The rearrangement of acrylate pathway genes to overcome the postulated metabolic imbalance and circumvent the bottleneck created by the Acr led to an increase in propionate production by almost four-fold. Interestingly, introduction of the optimized PSOs led to a shift of metabolic products from solvents to acids. While the non-induced C. saccharoperbutylacetonicum strain accumulated 83.8 mM acetate and 69.4 mM butyrate, the induced strain first produced 57.9 mM lactate, which was partially reassimilated and used for butyrate and propionate formation. The reason for this shift is not completely clear, however, can possibly be explained by the increased demand of the strain for ATP to maintain and express the enlarged PSOs. Both acetate and butyrate formation are important energy sources as they involve the formation of one ATP via substrate-level phosphorylation (Boynton et al. 1996; Hartmanis 1987), thus increasing the amount of available ATP. With an increased turnover of glucose to acids, the demand for reducing equivalents, especially oxidized ferredoxin but also NADH, increases in parallel as these are needed by the pyruvate:ferredoxin oxidoreductase, 3-hydroxybutyryl-CoA dehydrogenase, and butyryl-CoA dehydrogenase reactions (Jones and Woods 1986). In addition, the acrylate pathway consumes NADH during several steps. This raises the question of how the strain can adapt its metabolism to meet this increased need for reducing power. Under normal conditions, carbon flow from glucose to each of the acids and solvents happens in a particular ratio so that carbon and redox balances are closed (Jones and Woods 1986). However, solventogenesis is a process involving a high turnover of NADH to form ethanol and butanol (Jones and Woods 1986). Thus, a reduction of solventogenesis could save a substantial amount of NADH to be invested in acidogenesis including propionate formation via the acrylate pathway. Since C. saccharoperbutylacetonicum strains harboring optimized PSOs only produced little solvents (Fig. 5), it seems as if solventogenesis was indeed spared for the benefit of the energetically more favorable acidogenic pathways. To meet the increased need for oxidized ferredoxin, the strain can use its Rnf complex to recover oxidized ferredoxin and form NADH, which is accompanied by the generation of an ion gradient (Poehlein et al. 2017). This in turn can be used by the ATPase for further ATP formation and to fill up the ATP pool.
Although propionate production was successfully increased, the overall titer of 3 mM is still low. Considering the partial propionate reassimilation observed in both growth experiments, propionate titers could have been higher had it not been reduced to propanol. The formation of propanol is most probably due to the uptake of propionate by the acetoacetyl-CoA:acetate/butyrate CoA-transferase, which has a broad substrate spectrum including propionate (Hartmanis et al. 1984). The resulting propionyl-CoA can then be converted to propanol by aldehyde and alcohol dehydrogenases. Aside from propanol, other by-products such as solvents and butyrate limit the level of produced propionate as their formation requires both carbon and reducing equivalents. Other studies targeting heterologous propionate production using different host strains also reported challenges leading to mixed results. While E. coli engineered with the Sleeping beauty mutase operon and carrying multiple gene deletions or recombinant Ps. putida cultivated in fed-batch mode achieved a maximum of approx. 160 and 823 mM propionate from 326 mM glycerol and 850 mM L-threonine, respectively (Akawi et al. 2015; Mu et al. 2021), bacterial strains modified with the acrylate pathway also only produced rather low amounts of propionate. These ranged between 0.01 mM for L. plantarum (Balasubramanian and Subramanian 2019) and 3.7 mM for E. coli (Kandasamy et al. 2013), the latter of which is comparable to the propionate concentration produced by C. saccharoperbutylacetonicum [pMTL83151_PtcdB_L_PtcdB_AA][pMTL82251_PbgaL_LPTT]. The low propionate titers could be due to metabolic imbalances leading to the accumulation of pathway intermediates, redox deficiencies, and low activities of recombinant enzymes as previously hypothesized by other groups (Balasubramanian and Subramanian 2019; Kandasamy et al. 2013). Despite these hurdles, there are options that might lead to an improved propionate production using C. saccharoperbutylacetonicum through which it could possibly outperform recombinant E. coli and Ps. putida strains or at least reach the same production level. One such option is the introduction of PSOs into strains harboring tailored mutations to improve carbon and redox balances. Since carbon and redox equivalents are predominantly invested in C4-producing pathways in C. saccharoperbutylacetonicum, manipulations in these metabolic branches might be promising. A deletion of 3-hydroxybutyryl-CoA dehydrogenase, crotonase, or aldehyde and alcohol dehydrogenases in either C. saccharoperbutylacetonicum or its close relative C. acetobutylicum led to a reduction or complete abolishment of butyrate or solvent formation (Baur 2022; Cooksley et al. 2012; Lehmann and Lütke-Eversloh 2011). Simultaneously, saved reducing equivalents were used for lactate or ethanol formation (Baur 2022; Lehmann and Lütke-Eversloh 2011). If such a strain carried the reductive acrylate pathway, this might lead to increased propionate concentrations. Other options for modifications to manipulate the carbon and electron flow in favor of the acrylate pathway would be the deletion of the hydrogenase gene cluster or the global regulator Spo0A. The ferredoxin hydrogenase of C. saccharoperbutylacetonicum produces hydrogen while simultaneously oxidizing reduced ferredoxin (Dada et al. 2013). Thus, a deletion of the hydrogenase could save reduced ferredoxin to be turned over by the Rnf complex, which as mentioned before, can use this for NADH generation. Deletion of spo0A was shown to reduce both solventogenesis and sporulation in many clostridial strains including C. saccharoperbutylacetonicum (Atmadjaja et al. 2019; Harris et al. 2002; Schwarz et al. 2017). Since sporulation is also a major contributor to culture heterogeneity (Tracy et al. 2010), a combination of spo0A deletion with the PSOs could result in increased propionate titers. Another option to overcome culture heterogeneity would be chromosomal integration of the PSOs as this would lead to a plasmid-independent expression and thereby eliminate plasmid loss or instability as possible limiting factors. Whether such an optimized C. saccharoperbutylacetonicum strain could then outperform native propionate producers such as propionibacteria remains questionable as even higher propionate titers were achieved by the cultivation of these bacteria in bioreactors using more sophisticated approaches that thus far have never been employed using C. saccharoperbutylacetonicum. The highest ever reported propionate concentrations are 1 M using P. acidipropionici (Liu et al. 2016) and 1.2 to 1.8 M using P. freudenreichii (Chen et al. 2013; Feng et al. 2011), when strains were cultivated in fed-batch mode with high cell density or immobilized cells and glucose or hydrolyzed sugar cane molasses as substrates. These concentrations were achieved with productivities of 4.3 to 7.7 mM h−1, which is 54- to 96-fold higher than the productivity of C. saccharoperbutylacetonicum [pMTL83151_PtcdB_L_PtcdB_AA][pMTL82251_PbgaL_LPTT] (0.08 mM h−1). Nevertheless, it is imaginable that fed-batch or continuous cultivation of further optimized C. saccharoperbutylacetonicum could lead to another increase in propionate concentration and thus make it a strain that can very well compete but probably not outperform native producers with respect to the propionate concentration and productivity. Overall, C. saccharoperbutylacetonicum was successfully engineered as a propionate producer. Although propionate titer was rather low with 3 mM, different options for further strain engineering and cultivation are conceivable to increase the performance of recombinant C. saccharoperbutylacetonicum strains. Successfully engineered and improved strains could then possibly be considered for commercial propionate production using sustainable resources such as lignocellulosic hydrolysates.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author upon reasonable request.
References
Akawi L, Srirangan K, Liu X, Moo-Young M, Chou CP (2015) Engineering Escherichia coli for high-level production of propionate. J Ind Microbiol Biotechnol 42:1057–1072. https://doi.org/10.1007/s10295-015-1627-4
Atmadjaja AN, Holby V, Harding AJ, Krabben P, Smith HK, Jenkinson ER (2019) CRISPR-Cas, a highly effective tool for genome editing in Clostridium saccharoperbutylacetonicum N1–4(HMT). FEMS Microbiol Lett 366:fnz059. https://doi.org/10.1093/femsle/fnz059
Balasubramanian S, Subramanian R (2019) Metabolic perturbation of acrylate pathway in Lactobacillus plantarum. Biocatal Biotransformation 37:310–316. https://doi.org/10.1080/10242422.2019.1606215
Baur ST (2022) Construction of acid-producing Clostridium saccharoperbutylacetonicum strains by deletion, overexpression, and interfering with regulation of genes. Dissertation, University of Ulm. https://doi.org/10.18725/OPARU-42087
Beck MH (2020) Biological conversion of CO2 into the platform chemicals lactate and 3-hydroxypropionate using recombinant strains of Acetobacterium woodii. Dissertation, University of Ulm. https://doi.org/10.18725/OPARU-28743
Beck MH, Flaiz M, Bengelsdorf FR, Dürre P (2020) Induced heterologous expression of the arginine deiminase pathway promotes growth advantages in the strict anaerobe Acetobacterium woodii. Appl Microbiol Biotechnol 104:687–699. https://doi.org/10.1007/s00253-019-10248-9
Beck MH, Poehlein A, Bengelsdorf FR, Schiel-Bengelsdorf B, Daniel R, Dürre P (2016) Draft genome sequence of the strict anaerobe Clostridium neopropionicum X4 (DSM 3847T). Genome Announc 4:e00209-e216. https://doi.org/10.1128/genomeA.00209-16
Binder D, Probst C, Grünberger A, Hilgers F, Loeschcke A, Jaeger K-E, Kohlheyer D, Drepper T (2016) Comparative single-cell analysis of different E. coli expression systems during microfluidic cultivation. PLoS One 11:e0160711. https://doi.org/10.1371/journal.pone.0160711
Boynton ZL, Bennett GN, Rudolph FB (1996) Cloning, sequencing, and expression of genes encoding phosphotransacetylase and acetate kinase from Clostridium acetobutylicum ATCC 824. Appl Environ Microbiol 62:2758–2766. https://doi.org/10.1128/aem.62.8.2758-2766.1996
Chen F, Feng X, Xu H, Zhang D, Ouyang P (2013) Propionic acid production in a plant fibrous-bed bioreactor with immobilized Propionibacterium freudenreichii CCTCC M207015. J Biotechnol 164:202–210. https://doi.org/10.1016/j.jbiotec.2012.08.025
Cho C, Jang Y-S, Moon HG, Lee J, Lee SY (2015) Metabolic engineering of clostridia for the production of chemicals. Biofuel Bioprod Biorefin 9:211–225. https://doi.org/10.1002/bbb.1531
Cooksley CM, Zhang Y, Wang H, Redl S, Winzer K, Minton NP (2012) Targeted mutagenesis of the Clostridium acetobutylicum acetone-butanol-ethanol fermentation pathway. Metab Eng 14:630–641. https://doi.org/10.1016/j.ymben.2012.09.001
Dada O, Yusoff WMW, Kalil MS (2013) Biohydrogen production from ricebran using Clostridium saccharoperbutylacetonicum N1–4. Int J Hydrog Energy 38:15063–15073. https://doi.org/10.1016/j.ijhydene.2013.07.048
Dishisha T, Alvarez MT, Hatti-Kaul R (2012) Batch- and continuous propionic acid production from glycerol using free and immobilized cells of Propionibacterium acidipropionici. Bioresour Technol 118:553–562. https://doi.org/10.1016/j.biortech.2012.05.079
Fagan RP, Fairweather NF (2011) Clostridium difficile has two parallel and essential Sec secretion systems. J Biol Chem 286:27483–27493. https://doi.org/10.1074/jbc.m111.263889
Feng J, Zhang J, Ma Y, Feng Y, Wang S, Guo N, Wang H, Wang P, Jiménez-Bonilla P, Gu Y, Zhou J, Zhang Z-T, Cao M, Jiang D, Wang S, Liu X-W, Shao Z, Borovok I, Huang H, Wang Y (2021) Renewable fatty acid ester production in Clostridium. Nat Commun 12:4368. https://doi.org/10.1038/s41467-021-24038-3
Feng X, Chen F, Xu H, Wu B, Li H, Li S, Ouyang P (2011) Green and economical production of propionic acid by Propionibacterium freudenreichii CCTCC M207015 in plant fibrous-bed bioreactor. Bioresour Technol 102:6141–6146. https://doi.org/10.1016/j.biortech.2011.02.087
Flaiz M, Baur T, Gaibler J, Kröly C, Dürre P (2022) Establishment of green- and red-fluorescent reporter proteins based on the fluorescence-activating and absorption-shifting tag for use in acetogenic and solventogenic anaerobes. ACS Synth Biol 11:953–967. https://doi.org/10.1021/acssynbio.1c00554
Flaiz M, Ludwig G, Bengelsdorf FR, Dürre P (2021) Production of the biocommodities butanol and acetone from methanol with fluorescent FAST-tagged proteins using metabolically engineered strains of Eubacterium limosum. Biotechnol Biofuels 14:117. https://doi.org/10.1186/s13068-021-01966-2
Gonzalez-Garcia RA, McCubbin T, Navone L, Stowers C, Nielsen LK, Marcellin E (2017) Microbial propionic acid production. Fermentation 3:21. https://doi.org/10.3390/fermentation3020021
Gonzalez-Garcia RA, McCubbin T, Turner MS, Nielsen LK, Marcellin E (2020) Engineering Escherichia coli for propionic acid production through the Wood-Werkman cycle. Biotechnol Bioeng 117:167–183. https://doi.org/10.1002/bit.27182
Green MR, Sambrook J (2012) Molecular cloning: a laboratory manual. Cold Spring Harbor, New York, NY, USA
Grosse-Honebrink A, Little GT, Bean Z, Heldt D, Cornock RHM, Winzer K, Minton NP, Green E, Zhang Y (2021) Development of Clostridium saccharoperbutylacetonicum as a whole cell biocatalyst for production of chirally pure (R)-1,3-butanediol. Front Bioeng Biotechnol 9:659895. https://doi.org/10.3389/fbioe.2021.659895
Hanahan D (1983) Studies on transformation of Escherichia coli with plasmids. J Mol Biol 166:557–580. https://doi.org/10.1016/S0022-2836(83)80284-8
Harris LM, Welker NE, Papoutsakis ET (2002) Northern, morphological, and fermentation analysis of spo0A inactivation and overexpression in Clostridium acetobutylicum ATCC 824. J Bacteriol 184:3586–3597. https://doi.org/10.1128/jb.184.13.3586-3597.2002
Hartman AH, Liu H, Melville SB (2011) Construction and characterization of a lactose-inducible promoter system for controlled gene expression in Clostridium perfringens. Appl Environ Microbiol 77:471–478. https://doi.org/10.1128/AEM.01536-10
Hartmanis MGN (1987) Butyrate kinase from Clostridium acetobutylicum. J Biol Chem 262:617–621. https://doi.org/10.1016/S0021-9258(19)75828-1
Hartmanis MGN, Klason T, Gatenbeck S (1984) Uptake and activation of acetate and butyrate in Clostridium acetobutylicum. Appl Microbiol Biotechnol 20:66–71. https://doi.org/10.1007/BF00254648
Heap JT, Pennington OJ, Cartman ST, Minton NP (2009) A modular system for Clostridium shuttle plasmids. J Microbiol Methods 78:79–85. https://doi.org/10.1016/j.mimet.2009.05.004
Herrmann G, Selmer T, Jessen HJ, Gokarn RR, Selifonova O, Gort SJ, Buckel W (2005) Two beta-alanyl-CoA:ammonia lyases in Clostridium propionicum. FEBS J 272:813–821. https://doi.org/10.1111/j.1742-4658.2004.04518.x
Hetzel M, Brock M, Selmer T, Pierik AJ, Golding BT, Buckel W (2003) Acryloyl-CoA reductase from Clostridium propionicum. An enzyme complex of propionyl-CoA dehydrogenase and electron-transferring flavoprotein. Eur J Biochem 270:902–910. https://doi.org/10.1046/j.1432-1033.2003.03450.x
Hoffmeister S, Gerdom M, Bengelsdorf FR, Linder S, Flüchter S, Öztürk H, Blümke W, May A, Fischer R-J, Bahl H, Dürre P (2016) Acetone production with metabolically engineered strains of Acetobacterium woodii. Metab Eng 36:37–47. https://doi.org/10.1016/j.ymben.2016.03.001
Jiang L, Cui H, Zhu L, Hu Y, Xu X, Li S, Huang H (2015) Enhanced propionic acid production from whey lactose with immobilized Propionibacterium acidipropionici and the role of trehalose synthesis in acid tolerance. Green Chem 17:250–259. https://doi.org/10.1039/c4gc01256a
Jiménez-Bonilla P, Feng J, Wang S, Zhang J, Wang Y, Blersch D, de Bashan LE, Gaillard P, Guo L, Wang Y (2021) Identification and investigation of autolysin genes in Clostridium saccharoperbutylacetonicum strain N1–4 for enhanced biobutanol production. Appl Environ Microbiol 87:e02442-20. https://doi.org/10.1128/AEM.02442-20
Jones DT, Woods DR (1986) Acetone-butanol fermentation revisited. Microbiol Rev 50:484–524. https://doi.org/10.1128/mr.50.4.484-524.1986
Jones SW, Paredes CJ, Tracy B, Cheng N, Sillers R, Senger RS, Papoutsakis ET (2008) The transcriptional program underlying the physiology of clostridial sporulation. Genome Biol 9:R114. https://doi.org/10.1186/gb-2008-9-7-r114
Kandasamy V, Vaidyanathan H, Djurdjevic I, Jayamani E, Ramachandran KB, Buckel W, Jayaraman G, Ramalingam S (2013) Engineering Escherichia coli with acrylate pathway genes for propionic acid synthesis and its impact on mixed-acid fermentation. Appl Microbiol Biotechnol 97:1191–1200. https://doi.org/10.1007/s00253-012-4274-y
Kosaka T, Nakayama S, Nakaya K, Yoshino S, Furukawa K (2007) Characterization of the sol operon in butanol-hyperproducing Clostridium saccharoperbutylacetonicum strain N1–4 and its degeneration mechanism. Biosci Biotechnol Biochem 71:58–68. https://doi.org/10.1271/bbb.60370
Lehmann D, Lütke-Eversloh T (2011) Switching Clostridium acetobutylicum to an ethanol producer by disruption of the butyrate/butanol fermentative pathway. Metab Eng 13:464–473. https://doi.org/10.1016/j.ymben.2011.04.006
Li L, Eom H-J, Park J-M, Seo E, Ahn JE, Kim T-J, Kim JH, Han NS (2012) Characterization of the major dehydrogenase related to D-lactic acid synthesis in Leuconostoc mesenteroides subsp. mesenteroides ATCC 8293. Enzyme Microb Technol 51:274–279. https://doi.org/10.1016/j.enzmictec.2012.07.009
Liu Z, Ge Y, Xu J, Gao C, Ma C, Xu P (2016) Efficient production of propionic acid through high density culture with recycling cells of Propionibacterium acidipropionici. Bioresour Technol 216:856–861. https://doi.org/10.1016/j.biortech.2016.06.023
Ma C, Mu Q, Wang L, Shi Y, Zhu L, Zhang S, Xue Y, Tao Y, Ma Y, Yu B (2020) Bio-production of high-purity propionate by engineering L-threonine degradation pathway in Pseudomonas putida. Appl Microbiol Biotechnol 104:5303–5313. https://doi.org/10.1007/s00253-020-10619-7
Mani N, Dupuy B (2001) Regulation of toxin synthesis in Clostridium difficile by an alternative RNA polymerase sigma factor. Proc Natl Acad Sci USA 98:5844–5849. https://doi.org/10.1073/pnas.101126598
Martin-Verstraete I, Peltier J, Dupuy B (2016) The regulatory networks that control Clostridium difficile toxin synthesis. Toxins 8:153. https://doi.org/10.3390/toxins8050153
Moncrief JS, Barroso LA, Wilkins TD (1997) Positive regulation of Clostridium difficile toxins. Infect Immun 65:1105–1108. https://doi.org/10.1128/IAI.65.3.1105-1108.1997
Mu Q, Shi Y, Li R, Ma C, Tao Y, Yu B (2021) Production of propionate by a sequential fermentation-biotransformation process via L-threonine. J Agric Food Chem 69:13895–13903. https://doi.org/10.1021/acs.jafc.1c05248
Poehlein A, Montoya Solano JD, Flitsch SK, Krabben P, Winzer K, Reid SJ, Jones DT, Green E, Minton NP, Daniel R, Dürre P (2017) Microbial solvent formation revisited by comparative genome analysis. Biotechnol Biofuels 10:58. https://doi.org/10.1186/s13068-017-0742-z
Samel U-R, Kohler W, Gamer AO, Keuser U, Yang S-T, Jin Y, Lin M, Wang Z, Teles JH (2018) Propionic acid and derivatives. In: Ullmann´s Encyclopedia of Industrial Chemistry. Wiley-VCH, Weinheim, Germany. https://doi.org/10.1002/14356007.a22_223.pub4
Schölmerich MC, Katsyv A, Sung W, Mijic V, Wiechmann A, Kottenhahn P, Baker J, Minton NP, Müller V (2018) Regulation of lactate metabolism in the acetogenic bacterium Acetobacterium woodii. Environ Microbiol 20:4587–4595. https://doi.org/10.1111/1462-2920.14412
Schwarz KM, Grosse-Honebrink A, Derecka K, Rotta C, Zhang Y, Minton NP (2017) Towards improved butanol production through targeted genetic modification of Clostridium pasteurianum. Metab Eng 40:124–137. https://doi.org/10.1016/j.ymben.2017.01.009
Siegele DA, Hu JC (1997) Gene expression from plasmids containing the araBAD promoter at subsaturating inducer concentrations represents mixed populations. Proc Natl Acad Sci USA 94:8168–8172. https://doi.org/10.1073/pnas.94.15.8168
Singh V, Singh H, Das D (2019) Optimization of the medium composition for the improvement of hydrogen and butanol production using Clostridium saccharoperbutylacetonicum DSM 14923. Int J Hydrog Energy 44:26905–26919. https://doi.org/10.1016/j.ijhydene.2019.08.125
Stowers CC, Cox BM, Rodriguez BA (2014) Development of an industrializable fermentation process for propionic acid production. J Ind Microbiol Biotechnol 41:837–852. https://doi.org/10.1007/s10295-014-1423-6
Streett HE, Kalis KM, Papoutsakis ET (2019) A strongly fluorescing anaerobic reporter and protein-tagging system for Clostridium organisms based on the fluorescence-activating and absorption-shifting tag protein (FAST). Appl Environ Microbiol 85:e00622-e719. https://doi.org/10.1128/AEM.00622-19
Tracy BP, Gaida SM, Papoutsakis ET (2010) Flow cytometry for bacteria: enabling metabolic engineering, synthetic biology and the elucidation of complex phenotypes. Curr Opin Biotechnol 21:85–99. https://doi.org/10.1016/j.copbio.2010.02.006
Tracy BP, Jones SW, Fast AG, Indurthi DC, Papoutsakis ET (2012) Clostridia: the importance of their exceptional substrate and metabolite diversity for biofuel and biorefinery applications. Curr Opin Biotechnol 23:364–381. https://doi.org/10.1016/j.copbio.2011.10.008
Tummala SB, Welker NE, Papoutsakis ET (1999) Development and characterization of a gene expression reporter system for Clostridium acetobutylicum ATCC 824. Appl Environ Microbiol 65:3793–3799. https://doi.org/10.1128/AEM.65.9.3793-3799.1999
Wang P, Feng J, Guo L, Fasina O, Wang Y (2019) Engineering Clostridium saccharoperbutylacetonicum for high level isopropanol-butanol-ethanol (IBE) production from acetic acid pretreated switchgrass using the CRISPR-Cas9 system. ACS Sustain Chem Eng 7:18153–18164. https://doi.org/10.1021/acssuschemeng.9b05336
Wang Z, Ammar EM, Zhang A, Wang L, Lin M, Yang S-T (2015) Engineering Propionibacterium freudenreichii subsp. shermanii for enhanced propionic acid fermentation: effects of overexpressing propionyl-CoA:succinate CoA transferase. Metab Eng 27:46–56. https://doi.org/10.1016/j.ymben.2014.10.005
Weitz S, Hermann M, Linder S, Bengelsdorf FR, Takors R, Dürre P (2021) Isobutanol production by autotrophic acetogenic bacteria. Front Bioeng Biotechnol 9:657253. https://doi.org/10.3389/fbioe.2021.657253
Wirth S, Dürre P (2021) Investigation of putative genes for the production of medium-chained acids and alcohols in autotrophic acetogenic bacteria. Metab Eng 66:296–307. https://doi.org/10.1016/j.ymben.2021.04.010
Zhang L, Fan F, Palmer LM, Lonetto MA, Petit C, Voelker LL, St. John A, Bankosky B, Rosenberg M, McDevitt D (2000) Regulated gene expression in Staphylococcus aureus for identifying conditional lethal phenotypes and antibiotic mode of action. Gene 255:297–305. https://doi.org/10.1016/s0378-1119(00)00325-5
Acknowledgements
We kindly thank Dr. Maximilian Flaiz for providing plasmid pMTL83251_Ppta-ack_FAST as well as for the advice regarding the FAST assays. Furthermore, the help of Dr. Sebastian Reich regarding flow cytometry analysis is gratefully acknowledged.
Funding
Open Access funding enabled and organized by Projekt DEAL. This work was supported by the European Research Area Cofund on BioTechnologies (ERACoBioTech) within its project “Bioprocesses for the optimized, integrated production of butyl esters from sustainable resources” (BESTER; grants 161B0594A (DE) and 284538 (NO)).
Author information
Authors and Affiliations
Contributions
TB designed and performed the experiments, analyzed the data, and wrote the manuscript. AW acquired funding, participated in discussions, and revised the manuscript. PD supervised the work, revised the manuscript, and acquired funding. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose. The authors have no competing interests to declare that are relevant to the content of this article.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Baur, T., Wentzel, A. & Dürre, P. Production of propionate using metabolically engineered strains of Clostridium saccharoperbutylacetonicum. Appl Microbiol Biotechnol 106, 7547–7562 (2022). https://doi.org/10.1007/s00253-022-12210-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-022-12210-8