Abstract
To establish a rapid and specific antigen detection method for porcine circovirus type 2 (PCV2), monoclonal antibodies (mAbs) were produced against the PCV2 epidemic strains and a red latex microsphere immunochromatographic strip was established. A total of eight anti-PCV2b and four anti-PCV2d mAbs were produced, and seven mAbs were confirmed to react with PCV2a, PCV2b, and PCV2d strains using an immunoperoxidase monolayer assay. The results of micro-neutralization tests showed that the mAbs 2C8, 9H4, 10G7, 7B9, and 7C7 had good neutralizing activity, whereas the neutralizing activity of the mAbs 4B3, 4C9, 6H9, and 7E2 was lower than 50%. Three mAbs, 4B3, 7C7, and 9H4, and PCV2 pAb were selected for the establishment of a red latex microsphere immunochromatographic strip, and the combination of mAb 7C7 labeled with red latex microspheres and mAb 9H4 exhibited the greatest detection ability. The immunochromatographic strip had minimum detection limits of 102.5 TCID50/0.1 ml, 100.7 TCID50/0.1 ml, and 101.5 TCID50/0.1 ml for PCV2a/CL, PCV2b/MDJ, and PCV2d/LNHC, respectively. Furthermore, no cross-reactivity was found for African swine fever virus, classical swine fever virus, porcine respiratory and reproductive syndrome virus, porcine parvovirus, porcine pseudorabies virus, porcine circovirus type 1, transmissible gastroenteritis virus, porcine epidemic diarrhea virus, porcine rotavirus, or porcine deltacoronavirus using the immunochromatographic strip. Using PCR as a reference standard, the detection sensitivity, specificity, and overall coincidence rate of the immunochromatographic strip were 81.13%, 100%, and 90.00%. Additionally, the detection ability of the immunochromatographic strip was correlated with that of virus titration. The immunochromatographic strip was used to detect 183 clinical disease samples, and the average positive detection rate was 22.95%. In summary, this method has good sensitivity and specificity and is simple, convenient, and quick to operate. It has high application value for on-site diagnosis of PCV2 and virus quantification.
Key points
• A red latex microsphere immunochromatographic strip for PCV2 detection was developed.
• The method was not only simple to operate, but also takes less time.
• The method had good sensitivity and specificity.
Similar content being viewed by others
Introduction
Porcine circovirus type 2 (PCV2) belongs to the family Circoviridae (Allan et al. 1998; Finsterbusch and Mankertz 2009) and is a single-stranded circular DNA virus without an envelope (Crowther et al. 2003). It mainly contains five open reading frames: ORF1, ORF2, ORF3,ORF4, and ORF5 (Hamel et al. 1998; He et al. 2013; Liu et al. 2006, 2005; Lv et al. 2015; Mankertz et al. 2004, 1998; Meehan et al. 1998). ORF1 and ORF2 have been the most comprehensively researched and their functions are most certain. ORF1 encodes Rep and Rep′ proteins, which are related to replication of viral DNA (Cheung. 2003; Mankertz et al. 2003). ORF2 encodes the only structural protein of the virus, Cap protein, 60 of which are assembled into the icosahedral virus particle (Hamel et al. 1998; Khayat et al. 2011; Saha et al. 2012).
PCV2 is approximately 17 nm in diameter and is one of the smallest mammalian pathogens (Allan et al. 1998). Although the virus is small in size and simple in structure, its pathogenicity is complicated. PCV2 infection can be subclinical or apparent. The former has no obvious clinical symptoms and is mainly characterized by a decline in average daily weight gain. The latter includes post-weaning multi-systemic wasting syndrome (PMWS), respiratory system disease syndrome, reproductive disorders, and intestinal diseases. These syndromes are collectively referred to as PCV2-related diseases (PCVADs) and have serious impacts and cause significant pressure on the pig industry (Opriessnig et al. 2008; Segalés 2012).
Based on genetic evolution analysis of the entire PCV2 gene or the ORF2 sequence, PCV2 is currently divided into six genotypes, PCV2a–PCV2f (Bao et al. 2018; Liu et al. 2018), of which PCV2a, PCV2b, and PCV2d are the main genotypes (Guo et al. 2010; Xiao et al. 2015). Prior to 2003, PCV2a was the main epidemic genotype, whereas the PCV2b strain became the main epidemic strain at the beginning of 2004 (Segalés et al. 2013). Since 2006, PCV2d strains have gradually spread and have become epidemic in many countries (Kwon et al. 2017; Lv et al. 2020; Ramos et al. 2017). This shows that there is a significant genotype change in PCV2. With the widespread use of PCV2 vaccines, the detection rate of PCV2 antigens in pigs has been significantly reduced. However, due to the complexity of the disease and the uneven quality of vaccines, a large number of PCV2 infections still occur and can even indirectly lead to the death of pigs (Opriessnig et al. 2013; Seo et al. 2014). Given the extreme diversity of the current epidemic strains, it is particularly important to establish a detection method that can rapidly and accurately detect PCV2 of various genotypes.
Conventional detection methods for PCV2 antigen generally include polymerase chain reaction (PCR), quantitative PCR, virus isolation and identification, enzyme-linked immunosorbent assay (ELISA), chemiluminescent immunoassay (CLIA), and colloidal gold immunochromatographic assay (GICA). PCR-related detection methods have excellent detection sensitivity; however, such methods cannot distinguish between the genetic fragments of the virus and the infectious virus. The isolation and identification of PCV2 is the gold standard for the diagnosis of PCVAD. However, professional technician in a well-equipped laboratory is required and the detection process is long. Therefore, they are not suitable for routine on-site detection at all pig farms (Yin et al. 2020; Yu et al. 2018). Although the ELISA method is easier than virus isolation, it also requires professional equipment. Using of CLIA has gradually increased in the veterinary field in recent years because of its high detection efficiency and good specificity (Wang et al. 2020), but this method also requires professional equipment and is not suitable for large-scale field surveys. Compared with PCR, virus isolation and identification, ELISA and CLIA, GICA does not require professional technicians or equipment, and the detection time is much shorter. Most importantly, the results can be obtained by simple sampling and visual measurements (Siu et al. 2016; Yang et al. 2020b). Colored latex microspheres are a novel technology that has developed rapidly in recent years. Colored microsphere immunolabeling has many advantages, including visual results based on bright and rich colors, uniform particle size, good monodispersity, low prices, high reproducibility of test results, and convenient use. In particular, its sensitivity is higher than that of GICA. In addition, the color-rendering speed of this method is slightly faster. Because of these advantages, latex microsphere immune layer technology has been widely used in many applications, such as bacterial, viral, and parasite detection, in recent years (Deng et al. 2017; Kim et al. 2015). In the diagnosis of animal diseases, this method was used in the rapid diagnosis of foot-and-mouth disease (Reid et al. 2001). In SARS-CoV-2 detection, Chinese State Food and Drug Administration of China approved the launch of the 2019-nCoV antigen detection kit (latex method) on March 12, 2022. However, the use of this method to detect PCV2 has not been reported.
In this study, virus-like particles (VLPs) of the Cap protein from the PCV2b and PCV2d strains were expressed with a eukaryotic expression system and used as immunogens to produce PCV2 broad-spectrum specific mAbs. A red latex microsphere immunochromatographic strip based on broad-spectrum PCV2 mAbs was established and applied to rapid detection of PCV2. This method provides technical support for the rapid detection of the virus and is of great significance for the prevention and control of PCVADs.
Materials and methods
Cells, viruses, and mice for the elaboration of monoclonal antibodies
SP2/0 cells (ATCC® CRL-1581™) and PK15 cells (ATCC® CCL-33™) free of PCV1 and PCV2 were grown in Dulbecco’s modified eagle medium (DMEM, Invitrogen, Carlsbad, CA, USA) containing 10% heat-inactivated fetal bovine serum (FBS, Gibco, Grand Island, NY, USA), 100 U/mL penicillin, and 0.1 mg/mL streptomycin for mAb preparation and virus propagation. Different PCV2 strains and one PCV1/G strain were used in this study. Strain origins, genotypes, and accession numbers are shown in Table 1. The experimental animals used for mAb preparation were 6-week-old female BALB/c mice provided by the Weitong Lihua Laboratory Animal Technology Co., Ltd. (Beijing, China). MAb 5F2 (Huang et al. 2011), mAb 3A5 (Huang et al. 2019), and PCV2 polyclonal antibody (pAb) (Huang et al. 2019) were used as controls.
PCV2 challenged pigs
Six healthy 32-day-old pigs found to be negative for PCV2, African swine fever virus (ASFV), porcine reproductive and respiratory syndrome virus (PRRSV), and pseudorabies virus (PRV) by PCR detection were used for challenge tests. Pigs 91, 92, and 93 were designated as the mock-infected control group. Pigs 76, 78, and 79 were challenged intranasally with PCV2b/MDJ cultures (1.0 × 106.0 TCID50/ml) at a dose of 1 ml. Anal swabs were collected weekly.
Farm samples
One hundred samples, including 50 inguinal lymph nodes, lungs, and anal swab samples from PMWS-affected farms and 50 same kind samples from PCV2-negative pigs were collected for coincidence studies.
From 2018 to 2021, 183 lung or inguinal lymph node samples were collected from pigs with respiratory or wasting symptoms in 10 provinces of China.
Lung and inguinal lymph node samples were ground into a 0.1-g/ml emulsion in DMEM. Anal swabs were diluted with 500 μl of sample diluent and mixed well.
Production of mAbs
VLPs of the Cap protein of the PCV2b/MDJ and PCV2d/SDRS strains were expressed, purified, and emulsified with ISA15VG adjuvant (Seppic, Fairfield, NJ, USA) as described previously (Bian et al. 2021). Three 6-week-old BALB/c mice were immunized three times with each protein. For the first immunization, each mouse was immunized with 100 μg protein via multiple subcutaneous injections. A boost was administered 3 weeks later with the same immunization route and dose as the first immunization. Two weeks after the second immunization, blood was collected from the mandibular vein and an immunoperoxidase monolayer assay (IPMA) was used to detect serum antibody titers as previously described (Huang et al. 2011). When the antibody titer was more than 3200, a third immunization was given via intraperitoneal injection with a 1-ml dose containing 100-μg purified VLPs. Mice were euthanized 3 days later, and spleen cells were fused with SP2/0 cells as previously described (Huang et al. 2011). IPMA plates prepared with PCV2b/MDJ were used to identify positive hybridomas derived from PCV2b/MDJ VLPs immunized mice as described previously (Liu et al. 2004). Similarly, PCV2d-derived hybridomas were selected with IPMA plates prepared with PCV2d/SDRS. Positive hybridomas were cloned three times using the limiting dilution method, and the obtained positive hybridoma cells were frozen.
Incomplete Freund’s adjuvant was injected intraperitoneally into 6–7-week-old BALB/c mice at a dose of 0.5 ml/mouse, and approximately 1 × 106 hybridomas/mouse were intraperitoneally injected 1 week later. After 7 days, mouse ascites were collected and centrifuged at 3000 × g for 10 min. The supernatant was collected, aliquoted and stored at − 20 °C.
Identification of mAbs
Supernatants containing mAbs secreted by each positive hybridoma cell were collected, and their subclasses were determined using the SBA Clonotyping System-HRP antibody subclass detection kit (SouthernBiotech, Birmingham, AL, USA). PCV2a/CL, PCV2b/MDJ, and PCV2d/SDRS were used to prepare IPMA plates for antibody titration as described previously (Liu et al. 2004). MAb ascites were diluted two-fold from 1:1000 to 1:512,000, and then their IPMA antibody titers were tested as described previously (Huang et al. 2011) The results were observed under an optical microscope (Nikon Eclipse, TE2000-S, Tokyo, Japan) to determine the IPMA antibody titer of ascites against the three PCV2 genotypes. The reciprocal of the highest dilution at which the IPMA was positive was defined as the IPMA titer of the antibody.
The reactivity of the mAbs to the PCV2 was determined using Western blot as previously described with minor modifications (Huang et al. 2011). PCV2b/MDJ and PCV2d/SDRS cell cultures were used as detection antigens for the corresponding mAbs. The mAb 5F2 against PCV2-Cap (Huang et al. 2011) and the supernatant of SP2/0 cells were used as positive and negative controls, respectively. IRDye 800CW Goat anti-mouse IgG (H + L) was used as a secondary antibody for Western blot analysis, and the experimental results were visualized using an Infrared Imaging System (Odyssey CLx Studio 3.1, Gene Company Limited, USA).
To test the neutralizing activity of the mAbs, a sensitive neutralization assay was performed as previously described (Huang et al. 2019). Briefly, 300 μl of 104.3 TCID50 PCV2b/MDJ or PCV2d/SDRS was incubated for 1 h at 37 °C with an equal volume of undiluted hybridoma supernatant against PCV2b or PCV2d. After incubation, 400 μl of mixture was added evenly to four wells of a 96-well plate containing 50% confluent monolayers of PK15 cells and incubated at 37 °C for 1 h. After three washes with DMEM, 100 μl DMEM containing 2% FBS was added to each well. PK15 cells were fixed with cold methanol after incubation for 36 h at 37 °C with 5% CO2. IPMA was performed as described by Liu et al. (2004) with PCV2 pAb. The number of infected cells per well was determined by light microscopy (Nikon Eclipse), and the neutralizing activity of the antibody was calculated according to the following formula: neutralizing activity (%) = (number of positive cells in the negative control – number of positive cells in the antibody tested)/number of positive cells in the negative control × 100 (Huang et al. 2019). When the average neutralizing activity of the hybridoma supernatant was greater than 50%, the mAb was considered to have neutralizing activity. MAb 3A5 and mAb 5F2 (Huang et al. 2011, 2019) were used as positive and negative controls, respectively. Neutralizing activity assays were performed in triplicate for each mAb.
Eight stable anti-PCV2b hybridomas, named 1G5, 2C8, 4B3, 4C9, 6H9, 7E2, 9H4, and 10G7 and four stable anti-PCV2d hybridomas, named 2G8, 7B9, 7C7, and 7D1, were obtained after screening and cloning. Their isotypes were identified and mAbs 4B3, 4C9, 6H9, 10G7, and 7B9 were IgG1, 1G5, 7E2, 7C7, and 7D1 were IgG2a, 9H4, and 2G8 were IgG2b, 2C8, was IgG3, and the light chains of all 12 mAbs were kappa type.
Establishment of a red microsphere immunochromatographic strip
Three mAbs (4B3, 7C7, and 9H4) with broad-spectrum reactivity and high antibody titers together with PCV2 pAb were selected and purified using protein G Sepharose™ CL-4B (GE Healthcare, Uppsala, Sweden) according to the manufacturer’s instructions. MAb 4B3 and 9H4 were derived from PCV2b/MDJ while mAb 7C7 was derived from PCV2d/SDRS. Purified mAb 4B3, 7C7, and 9H4 were coupled respectively with red latex microspheres as described previously (Yu et al. 2011) and sprayed separately onto the glass fibers (Sigma-Aldrich, St. Louis, MO, USA) to create the binding pads. Purified mAbs 4B3, 7C7, 9H7, and PCV2 pAb were diluted to a final concentration of 1 μg/ml and fixed to the nitrocellulose (NC) membrane (Sartorius, Gottingen, Germany) as a detection line. Goat anti-mouse IgG (H + L) (Sigma-Aldrich) was diluted to a final concentration of 1 μg/ml and fixed to the NC membrane (Sartorius, Gottingen, Germany) as a quality control. Both detection and quality control lines were dried at 37 °C for 2 h. Binding pads coated with different labeled antibodies and NC reaction membranes coated with different antibodies were combined to make test strips. The positive and negative controls were diluted tenfold from 1:10 to 1:1000 with 0.05 M phosphate-buffered saline (PBS, pH 7.4) containing 0.5% NP-40 and tested using the strips. The optimal antibody combination was chosen based on the highest titer of the positive control.
MAb 7C7 coupled with red latex microspheres was sprayed onto the glass fiber (Sigma-Aldrich) at 15 μl/cm to create the binding pad and dried at 37 °C for 2 h. Then, purified mAb 9H4 and goat anti-mouse IgG were separately sprayed onto the NC membrane detection and control lines. The two lines were separated by 1 cm and dried at 37 °C for 2 h. Finally, the sample pad, binding pad, NC film, and absorbent pad were assembled onto a PVC-bottom plate, cut into 2.5-cm strips using a strip cutter and covered with plastic to create a PCV2 red microsphere immunochromatographic strip. To obtain the best test results, the optimal concentrations of coupled mAb 7C7, mAb 9H4, and goat anti-mouse IgG were investigated by varying concentrations for each antibody while maintaining concentrations for other antibodies constant.
Evaluation of the red microsphere immunochromatographic strip
Three PCV2 strains, PCV2a/CL (104.5 TCID50/0.1 ml), PCV2b/MDJ (104.7 TCID50/0.1 ml), and PCV2d/LNHC (105.5 TCID50/0.1 ml), were selected to test the sensitivity of the red latex microsphere immunochromatographic strip. Three viruses were serially diluted tenfold from 1:10 to 1:100,000 and the immunochromatographic strips were used to detect diluted samples in triplicate. The highest positive dilution was used to calculate the minimum virus detection amount (virus titer/highest positive dilution) in TCID50/0.1 ml.
To assess the detection range and cross-reactivity of the red latex microsphere immunochromatographic strip, eight different PCV2 strains, including PCV2a, PCV2b, and PCV2d; PCV1/G; porcine parvovirus type 1 (PPV1); PRV Bartha-K61 strain; PRRSV HN112 strain; classical swine fever virus (CSFV) C strain; ASFV, transmissible gastroenteritis virus (TGEV); porcine epidemic diarrhea virus (PEDV) porcine rotavirus (PRoV) (G5); and porcine deltacoronavirus (PDCoV) were tested in triplicate using the immunochromatographic strip.
To evaluate the sensitivity and specificity of the red latex microsphere immunochromatographic strip, the PCR method for detecting the PCV2 genome was used as the detection standard. The sample processing method was as follows: 100 samples from PMWS-affected farms and PCV2 negative pigs were diluted three-fold, then tested using the immunochromatographic strip. For PCR detection, DNA was extracted from the emulsions of the above samples using a blood/cell/tissue genomic DNA extraction kit (Tiangen, Beijing, China) and amplified with PCV2-primer-CR (5′-GGAAGCTTCAGTAATTTATTTCATATGGAA-3′) and PCR-primer-CR (5′-GGAAGCTTTTTTATCACTTCGTAATGGTT-3′) using a Premix Taq kit (TaKaRa, Kyoto, Japan). PCR products were analyzed by agarose gel electrophoresis. The sensitivity and specificity of the immunochromatographic strip were calculated using the following formulae: sensitivity = true positives × 100/(true positives + false negatives) and specificity = true negatives × 100/(true negatives + false positives). By comparing results obtained by the two methods, kappa coefficient value was used to evaluate the effectiveness of the test strip (kappa coefficient is calculated by SPSS software). For the interpretation of agreement of the kappa statistic, a value of 0.2 to 0.4 is considered fair, 0.4 to 0.6 is moderate, 0.6 to 0.8 is good, and 0.8 to 0.9 is very good (Altman 1990).
To further evaluate the detection ability of the immunochromatographic strip, 10 positive and 10 negative samples were selected for PCV2 titration. Twenty samples were centrifuged at 2000 rpm for 10 min and sterilized with 0.22-μm filters. Filtered samples were serially diluted tenfold with DMEM, and undiluted, 10−1, 10−2, 10−3, 10−4, and 10−5 samples were added to four wells containing a 50% monolayer of PK15 cells at 100 μl/well. At the same time, four normal cell controls were set up. The PK15 cells were incubated at 37 °C with 5% CO2 for 1 h. Samples were then replaced with DMEM containing 2% FBS and incubated for 72 h. IPMA was performed as described by Liu et al. (2004) with mouse serum against PCV2-Cap and HRP-labeled rabbit anti-mouse IgG (H + L) (Sigma-Aldrich) as the primary and secondary antibodies, respectively. Virus titrations of samples were calculated according to the Reed-Muench method.
Three batches of immunochromatographic strips were prepared using the optimized conditions, and an equal number of immunochromatographic strips was randomly selected from each batch for sensitivity and specificity tests. PCV2d/SDRS was diluted tenfold from 1:10 to 1:10,000 and tested with three batches of strips in triplicate. PCV1/G, PPV1, PRV, PRRSV, CSFV, ASFV, TGEV, PEDV, PRoV (G5), and PDCoV were tested with three batches of strips in triplicate. Results were used to evaluate the inter-batch and intra-batch repeatability of the immunochromatographic strips.
Statistical analysis
All data from the sensitive neutralization assay were represented as means with standard deviation (SD) of three independent experiments and visualized in the GraphPad Prism 5.0 software (GraphPad Software, CA, USA). Kappa coefficient is calculated by IBM SPSS Statistics 26.0 software.
Results
Production of mAbs against PCV2b and PCV2d Cap proteins
PCV2a/LG, PCV2b/MDJ, and PCV2d/SDRS were used to test the antibody titers and cross-reactivity of the 12 mAbs. The results are shown in Table 2. mAbs 2C8, 4B3, 4C9, 6H9, 7C7, 9H4, and 10G7 were found to react with all three viruses, with the highest titers being 8000, 128,000, 128,000, 64,000, 512,000, 64,000, and 64,000, respectively. The mAbs 1G5, 7B9, 7D1, and 7E2 had positive reactions with the PCV2b/MDJ and PCV2d/SDRS strains, with antibody titers reaching up to 128,000. The mAb 2G8 reacted only with the PCV2d strain, and the antibody titer was 16,000.
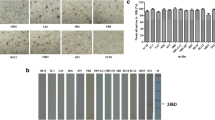
Western blot was used to assess the reaction characteristics of the 12 mAbs with the Cap protein of the corresponding PCV2 strain. Specific bands at approximately 28 kDa were detected for mAbs 4B3, 4C9, 6H9, 7E2, 10G7, 7B9, 7C7, and 7D1 and the positive control (Fig. 1a), but not for mAbs 1G5, 2C8, 9H4, or 2G8 or the negative control (Fig. 1a). We speculate that the mAbs 1G5, 2C8, 9H4, and 2G8 may recognize the conformation epitopes of the PCV2 Cap protein.
Characteristics of mAbs against PCV2. a Immunoreactivity analysis of mAbs by Western blotting. PCV2b/MDJ (oval dimension) and PCV2d/SDRS (triangle dimension) were separated by SDS-PAGE, transferred to nitrocellulose membranes, and then incubated with mAbs (as the primary antibody) and horseradish peroxidase (HRP)-labeled goat anti-mouse IgG (H + L) (1:4000, as the secondary antibody). MAb 5F2 and SP2/0 was used as positive and negative controls. Lane M is a protein molecular weight marker. b Neutralizing activity assays of mAbs for PCV2b/MDJ (mAb 1G5, 2C8, 4B3, 4C9, 6H9, 7E2, 9H4, and 10G8) or PCV2d/SDRS (mAb 2G8, 7B9, 7C7, 7D1, 3A5, and 5F2) by the sensitive neutralization assay. MAb 3A5 and 5F2 was used as positive and negative controls. The neutralizing activities of mAbs were expressed as the percentage reduction in the number of infected cells in comparison with negative control. A mean neutralizing activity of > 50% was considered to represent neutralization. Error bars represent the SD of neutralizing activities
Micro-neutralization tests were used to assess the ability of the 12 mAbs to neutralize PCV2b/MDJ or PCV2d/SDRS. The neutralizing activities of mAbs 2C8, 9H4, 10G7, 2G8, 7B9, 7C7, and 7D1 were found to be 80–100% (Fig. 1b), whereas the neutralizing activities of mAbs 1G5, 4B3, 4C9, 6H9, and 7E2 were less than 50% (Fig. 1b). Neutralizing activities of positive control mAb 3A5 and negative control mAb 5F2 were 86.05% and 2.65%.
Establishment of the red microsphere immunochromatographic strip
To establish a red microsphere immunochromatographic strip, mAb 4B3, 7C7, 9H4, and PCV2 pAb were purified and assessed by SDS-PAGE (Fig. 2). The results showed 55 kDa heavy chains and 20–25 kDa light chains among the purified mAbs or pAb (Fig. 2). There were few bands in other positions (Fig. 2), indicating that the purity of the purified antibodies was high and that they were suitable for establishment of the immunochromatographic strip.
To select the best pair of antibodies to establish a red latex microsphere immunochromatographic strip, red latex microsphere-labeled antibodies 4B3, 7C7, and 9H4 were paired with the mAbs 4B3, 7C7, and 9H4 and PCV2 pAb to prepare immunochromatographic strips for detection of positive and negative controls. The results showed that the combinations of red latex microsphere-labeled mAb 7C7 and coated mAb 9H4 and mAb 7C7 were significantly better than other combinations (Table 3). However, the immunochromatographic strips prepared using the combination of mAb 7C7 and mAb 7C7 labeled with red latex microspheres had a lighter C-limit color when the positive samples were diluted 10 and 100 times (results not shown). Therefore, the combination of mAb 9H4 and mAb 7C7 labeled with red latex microspheres was preferred.
To optimize the red latex microsphere immunochromatographic strip, optimal concentrations of mAb 7C7 labeled with red latex microspheres, mAb 9H4 and goat anti-mouse IgG were determined. The results showed that the optimal concentrations of labeled mAb 7C7, detection antibody 9H4, and the quality control goat anti-mouse IgG were 1.0 µg/ml, 0.5 µg/ml, and 0.5 µg/ml, respectively.
Validation of the red microsphere immunochromatographic strip
To test the sensitivity of the red latex microsphere immunochromatographic strip, PCV2a/CL (104.5 TCID50/0.1 ml), PCV2b/MDJ (104.7 TCID50/0.1 ml) and PCV2d/LNHC (105.5 TCID50/0.1 ml) were diluted tenfold and tested. The results showed that the titers of PCV2a/CL, PCV2b/MDJ, and PCV2d/LNHC were 100, 10,000, and 10,000, respectively (Fig. 3). The minimum detection limits of the immunochromatographic strip for PCV2a/CL, PCV2b/MDJ, and PCV2d/LNHC were calculated and found to be 102.5 TCID50/0.1 ml, 100.7 TCID50/0.1 ml, and 101.5 TCID50/0.1 ml, respectively. We conclude that the immunochromatographic strip was significantly more sensitive to the PCV2b and PCV2d strains than to PCV2a strains.
The specificity of the PCV2 red latex microsphere immunochromatographic strip to several porcine viruses was also tested, and the results showed that, with the exception of PCV2, all other viruses (PCV1, PPV, PRV, PRRSV, CSFV, AFSV, TGEV, PEDV, PRoV (G5), and PDCoV) were negative (Fig. 4). This demonstrated that the immunochromatographic strip specifically detected PCV2, and there was no cross-reactivity with other viruses.
One hundred samples were simultaneously tested by PCR and the red latex microsphere immunochromatographic strip, and the results are shown in Table 4. Using PCR, 53 samples were positive and 47 were negative, whereas 43 were positive and 57 were negative with the immunochromatographic strip. Of the 47 negative samples tested by PCR, all were negative with the immunochromatographic strip. However, 10 samples were negative with the immunochromatographic strip but positive with PCR. In total, 43 samples were positive and 47 samples were negative by both methods. The overall coincidence rate was 90.00% (90/100). Using PCR as the reference standard, the detection sensitivity of the immunochromatographic strip was 81.13% (43/53) and the specificity was 100% (47/47). This shows that the red latex microsphere immunochromatographic strip has excellent specificity and good sensitivity. The kappa coefficient for the comparison of this immunochromatographic strip method and the PCR method kit, was 0.720, suggesting that the strip could be used to detect PCV2.
Quantitative virus isolation was used to further evaluate the results of the red latex microsphere immunochromatographic strip. Ten positive samples and 10 negative samples tested by the immunochromatographic strip were selected for virus titration. The results are shown in Table 5. For samples that tested negative by the immunochromatographic strip, the virus titers were all less than the minimum detection limit of the virus titration, and the PCV2 titers of positive samples were between 101.7 and 105.3 TCID50/ml.
To evaluate the repeatability of immunochromatographic strip tests, three batches were used to repeatedly detect PCV2b/SDRS and PCV1/G, PPV1, PRV, PRRSV, CSFV, ASFV, TGEV, PEDV, PRoV (G5), and PDCoV. The results showed that the titer of PCV2b/SDRS determined using the immunochromatographic strip was 10,000 while other viruses were all negative for three batches. These results indicate that the method has good repeatability.
Detection of PCV2 in commercial farms
The PCV2 red latex microsphere immunochromatographic strip was used to assess a total of 183 samples of lung or inguinal lymph nodes from pigs with respiratory symptoms or wasting symptoms and feces from pigs with diarrhea collected between 2018 and 2021 in 10 Chinese provinces. The test results are shown in Table 6. Positive samples were detected in every province, with a positive rate ranging from 16.67 to 33.33%. In the past 4 years, the positive rate of PCV2 has been above 20.00%, and the total positive rate is 22.95%.
Detection of PCV2 from anal swabs of infected pigs
The PCV2 red latex microsphere immunochromatographic strip was used to assess anal swabs from PCV2-challenged pigs. The test results showed that 100.00% (3/3) of anal swabs from PCV2-challenged pigs were positive at 2 weeks post-infection (wpi), 33.33% (1/3) of anal swabs were positive at 3 wpi, and all anal swabs were negative at 4 wpi. The anal swabs of healthy control pigs were all negative through the experiment.
Discussion
PCV2 infection is ubiquitous around the world. Its associated diseases can be classified as subclinical or clinical. Decreased average daily gain with no evident clinical signs is found in subclinical disease, whereas PMWS, respiratory system disease syndrome, reproductive disorders, and intestinal diseases are the main clinical diseases associated with PCV2 infection. The amount of PCV2 in the organs of clinically infected pigs is significantly higher than that in subclinically infected pigs.
PCR and qPCR are commonly used for diagnosis of infectious diseases (Saiki et al. 1985; Shi and Yang 2021; Yang et al. 2020a). However, PCV2 nucleic acid fragments have been detected in many substances, including a porcine-derived commercial pepsin, some rotavirus vaccines, water samples, farm air, house flies, soil, and store-bought pork products (Blunt et al. 2011; Esona et al. 2014; Fenaux et al. 2004; Garcia et al. 2012; Kim et al. 2008; Li et al. 2010; Verreault et al. 2010). Furthermore, the PCV2 genome is difficult to degrade in vivo and in vitro (Wei et al. 2018), which may explain the ubiquitous presence of viral DNA. Thus, the detection of PCV2 genetic material does not necessarily mean that the sample contains complete, viable, and infectious PCV2 virions. Therefore, Wei et al. (2019) regarded virus isolation as the gold standard for the diagnosis of PCV2-related diseases. Only when PCV2 is successfully isolated, or at least the full-length genome is amplified, and corresponding clinical symptoms and pathological changes are observed can PCV2 be considered responsible for a disease. However, the technical requirements for virus isolation are relatively high and it is a time consuming process, so it is necessary to develop a simple method to detect PCV2 virions instead of virus genes. Based on this, we used two broad-spectrum PCV2 mAbs to establish a red latex microsphere immunochromatographic strip for rapid detection of PCV2.
There have been many reports of the production of PCV2 mAbs and their application in PCV2 detection. As early as 2002, McNeilly et al. reported a capture ELISA method to detect purified PCV2 in PK15 cultures based on mAbs. In recent years, Huang et al. (2019) developed a semi-quantitative capture ELISA based on broad-spectrum mAb 3A5 derived from the PCV2a/LG strain for detection of PCV2 in cell cultures and found that the minimum detectable amount of the PCV2a/LG strain was 200 TCID50. However, the detection sensitivity of this ELISA is significantly better for the PCV2a strains than for the currently dominant PCV2b and PCV2d strains. Therefore, we prepared mAbs against PCV2b and PCV2d Cap proteins to improve detection sensitivity for the dominant strains.
Of the 12 mAbs prepared in this study, eight were positive for PCV2a/LG, PCV2b/MDJ, and PCV2d/LNHC. Among them, mAbs 2C8, 9H4, 10G7, and 7C7 had the ability to neutralize PCV2 (neutralizing activity > 88%), whereas the neutralizing activities of 4B3, 4C9, and 6H9 were less than 50%. Thus, we infer that the epitopes of these mAbs can be divided into at least two types: neutralizing epitopes and non-neutralizing epitopes. Among the neutralizing mAbs, the mAbs 2C8 and 9H4 reacted negatively with the PCV2-Cap protein, whereas mAb 10G7 and 7C7 reacted positively with the PCV2-Cap protein in Western blots. We conclude that the neutralizing epitopes of mAbs 2C8 and 9H4 were different from those of mAbs 10G7 and 7C7. MAbs 4B3, 9H4, and 7C7, which have different epitopes, and PCV2 pAb were selected for antibody pairing with the red latex microsphere immunochromatographic strip, and mAbs 9H4 and 7C7 were selected as the coating and labeled antibodies, respectively.
The red latex microsphere immunochromatographic strip based on mAbs 9H4 and 7C7 with broad-spectrum reactivity to the PCV2a, PCV2b, and PCV2d strains was confirmed to have outstanding specificity and sensitivity. The immunochromatographic strip detected only PCV2, including the PCV2a, PCV2b, and PCV2d strains, and exhibited no cross-reactivity with other common viruses. Regarding the sensitivity of the test strip, the minimum detection limit was significantly higher for the PCV2a/CL strain than for the PCV2b/MDJ and PCV2d/LNHC strains. The possible reason for this is that mAbs 9H4 and 7C7 were derived from the PCV2b and PCV2d strains, so their sensitivity would be higher for these two strains. This also reflects the fact that PCV2 genotype changes have caused differences in antigenicity.
PCR was used as the detection standard to evaluate the immunochromatographic strip, and it was found that the immunochromatographic strip had a detection sensitivity of 81.13% and a specificity of 100%. The total coincidence rate of the two methods was 90.00%. However, the detection sensitivity of the immunochromatographic strip was lower than that of PCR. The main reasons for this include the following: (1) PCR was used to detect PCV2 nucleic acids, whereas the immunochromatographic strip was used to detect PCV2 virus particles, and the presence of nucleic acids does not necessarily represent the presence of a complete virus; and (2) PCR is the process of nucleic acid amplification, and its sensitivity is recognized to be higher than that of general capture ELISA and immune layer detection strips. In addition, virus titration was also used to evaluate the immunochromatographic strip, and it was found that the test results of the two methods were consistent for 20 samples (Table 5). These results indicate that the immunochromatographic strip is suitable for the detection of PCV2 virus particles.
Based on the advantages and disadvantages of the immunochromatographic strip, it was suitable for the rapid diagnosis of PCVAD. If the strip and PCR test results are inconsistent, it should be comprehensively determined by virus isolation, clinical symptoms, histopathological lesions, and PCV2 amount in related tissues. In addition to the clinical symptoms and pathological damage, the diagnostic points of PCVAD (including PMWS, respiratory system disease syndrome, reproductive disorders, and intestinal diseases) also include that moderate to high amount of PCV2 are detected in damaged tissues (Segalés 2012). Furthermore, because the antibodies used in this method cannot discriminate PCV2 genotypes, further gene amplification and sequencing are required to determine the genotype of PCV2. Additional, the immunochromatographic strip could not achieve quantitative detection, but PCV2 amount of the tested sample could be indirectly estimated by parallel detection of serially diluted positive controls of known virus titer. It is worth mentioning that the virus could be successfully isolated if the sample was tested positive by this strip.
In this study, a series of PCV2 mAbs were prepared using epidemic strains as immunogens. Two broad-spectrum neutralizing mAbs were used to establish a red latex microsphere immunochromatographic strip for detecting PCV2 virus particles. This detection method can be used to detect strains from the three main PCV2 genotypes and displays no cross-reactivity with other common viruses. This method overcomes the shortcomings of other antigen detection methods, which are time consuming and require highly specialized equipment and personnel, as the immunochromatographic strip require only 10 min to visually observe the results. This detection method can be used for the rapid detection of virus in infected cell cultures and anal swabs and organ tissues of infected pigs and provides a methodological basis for the rapid diagnosis of PCV2-related diseases, determination of virus titers in tissue culture and quality control of vaccines.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Allan GM, McNeilly F, Kennedy S, Daft B, Clarke EG, Ellis JA, Haines DM, Meehan BM, Adair BM (1998) Isolation of porcine circoviruslike viruses from pigs with a wasting disease in the USA and Europe. J Vet Diagn Invest 10(1):3–10
Altman DG (1990) Practical statistics for medical research. Chapman and Hall, London, United Kingdom
Bao F, Mi S, Luo Q, Guo H, Tu C, Zhu G, Gong W (2018) Retrospective study of porcine circovirus type 2 infection reveals a novel genotype PCV2f. Transbound Emerg Dis 65:432–440
Bian HQ, Huang LP, Wei YW, Xia DL, Zhu HZ, Wu HL, Yu C, Liu JH, Feng L, Liu CM (2021) Preparation of porcine circovirus type 2 virus-like particles and its immunogenicity research. Chin Vet Sci 51:1007–1014 (in Chinese)
Blunt R, McOrist S, McKillen J, McNair I, Jiang T, Mellits K (2011) House fly vector for porcine circovirus 2b on commercial pig farms. Vet Microbiol 149:452–455
Cheung AK (2003) Transcriptional analysis of porcine circovirus type 2. Virology 305:168–180
Crowther RA, Berriman JA, Curran WL, Allan GM, Todd D (2003) Comparison of the structures of three circoviruses: chicken anemia virus, porcine circovirus type 2, and beak and feather disease virus. J Virol 77:13036–13041
Deng J, Liu Y, Jia R, Wang M, Chen S, Zhu D, Liu M, Sun K, Zhao X, Yin Z, Chen A (2017) Development of an immunochromatographic strip for detection of antibodies against duck Tembusu virus. J Virol Methods 249:137–142
Esona MD, Mijatovic-Rustempasic S, Yen C, Parashar UD, Gentsch JR, Bowen MD, LaRussa P (2014) Detection of PCV-2 DNA in stool samples from infants vaccinated with RotaTeq®. Hum Vaccin Immunother 10:25–32
Fenaux M, Opriessnig T, Halbur PG, Xu Y, Potts B, Meng XJ (2004) Detection and in vitro and in vivo characterization of porcine circovirus DNA from a porcine-derived commercial pepsin product. J Gen Virol 85:3377–3382
Finsterbusch T, Mankertz A (2009) Porcine circoviruses–small but powerful. Virus Res 143:177–183
Garcia LA, Viancelli A, Rigotto C, Pilotto MR, Esteves PA, Kunz A, Barardi CR (2012) Surveillance of human and swine adenovirus, human norovirus and swine circovirus in water samples in Santa Catarina, Brazil. J Water Health 10:445–452
Guo LJ, Lu YH, Wei YW, Huang LP, Liu CM (2010) Porcine circovirus type 2 (PCV2): genetic variation and newly emerging genotypes in China. Virol J 7:273
Hamel AL, Lin LL, Nayar GP (1998) Nucleotide sequence of porcine circovirus associated with postweaning multisystemic wasting syndrome in pigs. J Virol 72:5262–5267
He J, Cao J, Zhou N, Jin Y, Wu J, Zhou J (2013) Identification and functional analysis of the novel ORF4 protein encoded by porcine circovirus type 2. J Virol 87:1420–1429
Huang L, Lu Y, Wei Y, Guo L, Liu C (2011) Development of a blocking ELISA for detection of serum neutralizing antibodies against porcine circovirus type 2. J Virol Methods 171:26–33
Huang L, Wei Y, Xia D, Liu D, Zhu H, Wu H, Feng L, Liu C (2019) A broad spectrum monoclonal antibody against porcine circovirus type 2 for antigen and antibody detection. Appl Microbiol Biotechnol 103:3453–3464
Huang L, Sun Z, Xia D, Wei Y, Sun E, Liu C, Zhu H, Bian H, Wu H, Feng L, Wang J, Liu C (2020) Neutralization mechanism of a monoclonal antibody targeting a porcine circovirus type 2 Cap protein conformational epitope. J Virol 94.
Khayat R, Brunn N, Speir JA, Hardham JM, Ankenbauer RG, Schneemann A, Johnson JE (2011) The 2.3-angstrom structure of porcine circovirus 2. J Virol 85:7856–7862
Kim KH, Chang HW, Nam YD, Roh SW, Kim MS, Sung Y, Jeon CO, Oh HM, Bae JW (2008) Amplification of uncultured single-stranded DNA viruses from rice paddy soil. Appl Environ Microbiol 74:5975–5985
Kim YK, Lim SI, Cho IS, Cheong KM, Lee EJ, Lee SO, Kim JB, Kim JH, Jeong DS, An BH, An DJ (2015) A novel diagnostic approach to detecting porcine epidemic diarrhea virus: the lateral immunochromatography assay. J Virol Methods 225:4–8
Kwon T, Lee DU, Yoo SJ, Je SH, Shin JY, Lyoo YS (2017) Genotypic diversity of porcine circovirus type 2 (PCV2) and genotype shift to PCV2d in Korean pig population. Virus Res 228:24–29
Li L, Kapoor A, Slikas B, Bamidele OS, Wang C, Shaukat S, Masroor MA, Wilson ML, Ndjango JB, Peeters M, Gross-Camp ND, Muller MN, Hahn BH, Wolfe ND, Triki H, Bartkus J, Zaidi SZ, Delwart E (2010) Multiple diverse circoviruses infect farm animals and are commonly found in human and chimpanzee feces. J Virol 84:1674–1682
Liu C, Ihara T, Nunoya T, Ueda S (2004) Development of an ELISA based on the baculovirus-expressed capsid protein of porcine circovirus type 2 as antigen. J Vet Med Sci 66:237–242
Liu J, Chen I, Kwang J (2005) Characterization of a previously unidentified viral protein in porcine circovirus type 2-infected cells and its role in virus-induced apoptosis. J Virol 79:8262–8274
Liu J, Chen I, Du Q, Chua H, Kwang J (2006) The ORF3 protein of porcine circovirus type 2 is involved in viral pathogenesis in vivo. J Virol 80:5065–5073
Liu J, Wei C, Dai A, Lin Z, Fan K, Fan J, Liu J, Luo M, Yang X (2018) Detection of PCV2e strains in Southeast China. PeerJ 6:e4476
Lv Q, Guo K, Xu H, Wang T, Zhang Y (2015) Correction: identification of putative ORF5 protein of porcine circovirus type 2 and functional analysis of GFP-fused ORF5 protein. PLoS One 10:e0134203
Lv N, Zhu L, Li W, Li Z, Qian Q, Zhang T, Liu L, Hong J, Zheng X, Wang Y, Zhang Y, Chai J (2020) Molecular epidemiology and genetic variation analyses of porcine circovirus type 2 isolated from Yunnan Province in China from 2016–2019. BMC Vet Res 16:96
Mankertz J, Buhk HJ, Blaess G, Mankertz A (1998) Transcription analysis of porcine circovirus (PCV). Virus Genes 16:267–276
Mankertz A, Mueller B, Steinfeldt T, Schmitt C, Finsterbusch T (2003) New reporter gene-based replication assay reveals exchangeability of replication factors of porcine circovirus types 1 and 2. J Virol 77:9885–9893
Mankertz A, Caliskan R, Hattermann K, Hillenbrand B, Kurzendoerfer P, Mueller B, Schmitt C, Steinfeldt T, Finsterbusch T (2004) Molecular biology of Porcine circovirus: analyses of gene expression and viral replication. Vet Microbiol 98:81–88
McNeilly F, McNair I, O’Connor M, Brockbank S, Gilpin D, Lasagna C, Boriosi G, Meehan B, Ellis J, Krakowka S, Allan GM (2002) Evaluation of a porcine circovirus type 2-specific antigen-capture enzyme-linked immunosorbent assay for the diagnosis of postweaning multisystemic wasting syndrome in pigs: comparison with virus isolation, immunohistochemistry, and the polymerase chain reaction. J Vet Diagn Invest 14:106–112
Meehan BM, McNeilly F, Todd D, Kennedy S, Jewhurst VA, Ellis JA, Hassard LE, Clark EG, Haines DM, Allan GM (1998) Characterization of novel circovirus DNAs associated with wasting syndromes in pigs. J Gen Virol 79(Pt 9):2171–2179
Opriessnig T, Madson DM, Prickett JR, Kuhar D, Lunney JK, Elsener J, Halbur PG (2008) Effect of porcine circovirus type 2 (PCV2) vaccination on porcine reproductive and respiratory syndrome virus (PRRSV) and PCV2 coinfection. Vet Microbiol 131:103–114
Opriessnig T, Xiao CT, Gerber PF, Halbur PG (2013) Emergence of a novel mutant PCV2b variant associated with clinical PCVAD in two vaccinated pig farms in the U.S. concurrently infected with PPV2. Vet Microbiol 163:177–183
Ramos N, Porley D, Mirazo S, Castro G, Cabrera K, Lozano A, Arbiza J (2017) Molecular study of Porcine Circovirus type 2 in wild boars and domestic pigs in Uruguay from 2010 to 2014: Predominance of recombinant circulating strains. Gene 637:230–238
Reid S, Ferris N, Brüning A, Hutching G, Kowalska Z, Akerblom L (2001) Development of a rapid chromatographic strip test for the pen-side detection of foot-and-mouth disease virus antigen. J Virol Methods 96(2):189–202
Saha D, Huang L, Bussalleu E, Lefebvre DJ, Fort M, Van Doorsselaere J, Nauwynck HJ (2012) Antigenic subtyping and epitopes’ competition analysis of porcine circovirus type 2 using monoclonal antibodies. Vet Microbiol 157:13–22
Saiki RK, Scharf S, Faloona F, Mullis KB, Horn GT, Erlich HA, Arnheim N (1985) Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science 230:1350–1354
Segalés J (2012) Porcine circovirus type 2 (PCV2) infections: clinical signs, pathology and laboratory diagnosis. Virus Res 164:10–19
Segalés J, Kekarainen T, Cortey M (2013) The natural history of porcine circovirus type 2: from an inoffensive virus to a devastating swine disease? Vet Microbiol 165:13–20
Seo HW, Park C, Kang I, Choi K, Jeong J, Park SJ, Chae C (2014) Genetic and antigenic characterization of a newly emerging porcine circovirus type 2b mutant first isolated in cases of vaccine failure in Korea. Arch Virol 159:3107–3111
Shi MY, Yang FL (2021) RPA and its application in detection of animal pathogens. China Anim Husb Vet Med 48:467–476 (in Chinese)
Siu LK, Tsai YK, Lin JC, Chen TL, Fung CP, Chang FY (2016) Development of a colloidal gold-based immunochromatographic strip for rapid detection of Klebsiella pneumoniae serotypes K1 and K2. J Clin Microbiol 54:3018–3021
Verreault D, Létourneau V, Gendron L, Massé D, Gagnon CA, Duchaine C (2010) Airborne porcine circovirus in Canadian swine confinement buildings. Vet Microbiol 141:224–230
Wang F, Li ZF, Yang YY, Wan DB, Vasylieva N, Zhang YQ, Cai J, Wang H, Shen YD, Xu ZL, Hammock BD (2020) Chemiluminescent enzyme immunoassay and bioluminescent enzyme immunoassay for tenuazonic acid mycotoxin by exploitation of nanobody and nanobody-nanoluciferase fusion. Anal Chem 92:11935–11942
Wei R, Trus I, Yang B, Huang L, Nauwynck HJ (2018) Breed Differences in PCV2 uptake and disintegration in porcine monocytes. Viruses 10.
Wei R, Xie J, Theuns S, Nauwynck HJ (2019) Changes on the viral capsid surface during the evolution of porcine circovirus type 2 (PCV2) from 2009 till 2018 may lead to a better receptor binding. Virus Evol 5, vez026.
Xiao CT, Halbur PG, Opriessnig T (2015) Global molecular genetic analysis of porcine circovirus type 2 (PCV2) sequences confirms the presence of four main PCV2 genotypes and reveals a rapid increase of PCV2d. J Gen Virol 96:1830–1841
Yang DM, Chang TJ, Wang ML, Tsai PH, Lin TH, Wang CT, Liang KH (2020) Hunting severe acute respiratory syndrome coronavirus 2 (2019 novel coronavirus): from laboratory testing back to basic research. J Chin Med Assoc 83:524–526
Yang F, Xiao Y, Chen B, Wang L, Liu F, Yao H, Wu N, Wu H (2020) Development of a colloidal gold-based immunochromatographic strip test using two monoclonal antibodies to detect H7N9 avian influenza virus. Virus Genes 56:396–400
Yin M, Hu X, Sun Y, Xing Y, Xing G, Wang Y, Li Q, Wang Y, Deng R, Zhang G (2020) Broad-spectrum detection of zeranol and its analogues by a colloidal gold-based lateral flow immunochromatographic assay in milk. Food Chem 321:126697
Yu LL, Ding JZ, Wen LY, Lou D, Yan XL, Lin LJ, Lu SH, Lin DD, Zhou XN (2011) Development of a rapid dipstick with latex immunochromatographic assay (DLIA) for diagnosis of schistosomiasis japonica. Parasit Vectors 4:157
Yu X, Wei L, Chen H, Niu X, Dou Y, Yang J, Wang Z, Tang Y, Diao Y (2018) Development of colloidal gold-based immunochromatographic assay for rapid detection of goose parvovirus. Front Microbiol 9:953
Funding
This study was supported by the National Natural Science Foundation of China (grant no. 31873012).
Author information
Authors and Affiliations
Contributions
LH, CL, and LF conceived and designed research. CY and LH conducted experiments. YW and HZ contributed new reagents or analytical tools. CY and JH analyzed data. LH and CY wrote the manuscript. All authors read and approved the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Ethics approval
The experimental protocol was approved by the Experimentation and Laboratory Animal Welfare Committee of the Harbin Veterinary Research Institute (HVRI) of the Chinese Academy of Agricultural Sciences (CAAS) with the approval code of 201230–01 and 210512–04. All procedures involving animals were carried out in strict accordance with the recommendations in the guide for the care and use of laboratory animals of the Ministry of Science and Technology of the People’s Republic of China.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yu, C., Wei, Y., Zhang, H. et al. Rapid detection of porcine circovirus type 2 by a red latex microsphere immunochromatographic strip. Appl Microbiol Biotechnol 106, 5757–5769 (2022). https://doi.org/10.1007/s00253-022-12074-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-022-12074-y