Abstract
Target proteins in biotechnological applications are highly diverse. Therefore, versatile flexible expression systems for their functional overproduction are required. In order to find the right heterologous gene expression strategy, suitable host-vector systems, which combine different genetic circuits, are useful. In this study, we designed a novel Bacillus subtilis expression toolbox, which allows the overproduction and secretion of potentially toxic enzymes. This toolbox comprises a set of 60 expression vectors, which combine two promoter variants, four strong secretion signals, a translation-enhancing downstream box, and three plasmid backbones. This B. subtilis toolbox is based on a tailor-made, clean deletion mutant strain, which is protease and sporulation deficient and exhibits reduced autolysis and secondary metabolism. The appropriateness of this alternative expression platform was tested for the overproduction of two difficult-to-produce eukaryotic model proteins. These included the sulfhydryl oxidase Sox from Saccharomyces cerevisiae, which forms reactive hydrogen peroxide and undesired cross-linking of functional proteins, and the human interleukin-1β, a pro-inflammatory cytokine. For the best performing Sox and interleukin, overproducing and secreting variants of these new B. subtilis toolbox fermentation strategies were developed and tested. This study demonstrates the suitability of the prokaryotic B. subtilis host-vector system for the extracellular production of two eukaryotic proteins with biotechnological relevance.
Key points
• Construction of a versatile Bacillus subtilis gene expression toolbox.
• Verification of the toolbox by the secretory overproduction of two difficult-to-express proteins.
• Fermentation strategy for an acetoin-controlled overproduction of heterologous proteins.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Engineered Bacillus strains, such as Bacillus subtilis, are important workhorses for the controlled overproduction of target proteins. Bacillus subtilis exhibits two major advantages over the widely used Escherichia coli expression systems. Firstly, it is a GRAS (generally recognized as safe) organism and as an endotoxin-free platform highly suitable for applications especially in the food and feed industry (Yang et al. 2016; Taguchi et al. 2015) or as vaccine biofactory (Rosales-Mendoza et al. 2016). Secondly, it enables extracellular protein production (Schallmey et al. 2004; Harwood and Cranenburgh 2008) and is considered to be a supersecreting cell factory (van Dijl and Hecker 2013). However, a drawback of available B. subtilis expression strains, such as B. subtilis WB600 or B. subtilis WB800 (Wu et al. 1991, 2002), is the fact that they contain several antibiotic resistance genes, which are not desired in many applications (Emond et al. 2001). Hence, aided by techniques for genomic modifications, such as the Cre-loxP technique (Kumpfmüller et al. 2013), CRISPR-Cas9 (Zhang et al. 2018), or counter-selection systems (Liu et al. 2008; Yu et al. 2010), marker-free expression strains can be generated that are favorable for various applications. Recently, new markerless B. subtilis expression strains were developed, which exhibited improved metabolic or protein production activities (Liu et al. 2018a, 2018b; Fan et al. 2018). Such optimized B. subtilis expression hosts comprise novel, tailor-made strains with reduced extracellular proteolytic activities (Zhang et al. 2018; Zhao et al. 2019). Furthermore, genome-reduced strains, like B. subtilis PG10, have been shown to be improved cell factories, which enable a more efficient production of “difficult-to-produce proteins” (Aguilar Suárez et al. 2019) or lantibiotics (van Tilburg et al. 2020).
Each target protein is unique and demands special expression systems or conditions. Many heterologous proteins, especially those that originate from eukaryotic organisms, are frequently difficult-to-express. Furthermore, target proteins could be harmful to the host and can therefore not be overproduced in established expression hosts. For example, sulfhydryl oxidases (Sox) are promising enzymes for food applications to improve rheological properties of doughs for pasta and pastries (Faccio et al. 2011). However, Sox is difficult-to-produce with classical microbial expression systems due to its toxicity caused by hydrogen peroxide formation. Furthermore, Sox is responsible for undesired cross-linking of functional proteins. One means to overcome this problem is to use expression systems which are specifically designed for the production of such toxic enzymes (Zemella et al. 2015).
To date, a permanently increasing number of novel Bacillus host-vector systems is available. One recent example for B. subtilis is the LIKE system (Toymentseva et al. 2012), which is based on a strictly controlled promoter that is strongly induced by cell wall-specific antibiotics. Another example is the BioBrick toolbox, which comprises essential genetic building blocks like integrative and replicative expression vectors, different promoters, or epitope tags, and thus enables a standardized protein overproduction with B. subtilis (Radeck et al. 2013; Popp et al. 2017). However, none of the existing systems combines satisfyingly all required parameters for an efficient expression platform, which should include (i) a suitable vector backbone, (ii) a selection marker, (iii) a strictly controlled promotor, (iv) a strong terminator, and (v) a suitable host chassis. Furthermore, strong secretion signals and translation enhancer sequences might be beneficial. Due to this variety of parameters, a simple handling and the possibility to easily exchange specific genetic elements should be a prerequisite in the development of a suitable and well-structured expression platform.
In this study, we constructed an alternative expression platform for B. subtilis and designed a novel host-vector system for recombinant protein production, which tries to meet the abovementioned parameters and enables the overproduction of potential toxic target proteins. The new markerless B. subtilis expression strain is deficient in its eight main extracellular proteases and shows reduced lysis rates by deletion of the lytC gene (Kabisch et al. 2013) and no sporulation by a spoIIGA mutation. In addition, this prototrophic B. subtilis strain was further adapted by deletions in three major secondary metabolite gene clusters in order to prevent interfering background metabolic activities and to support the protein downstream processing. We demonstrate that this new B. subtilis strain with the toolbox of 60 different expression vectors enables the development of successful strategies for the secretory overproduction of sophisticated eukaryotic proteins like the yeast sulfhydryl oxidase Sox and the human growth factor interleukin-1β.
Materials and methods
Unless stated otherwise all chemicals used in this study were of analytical grade purchased from Carl Roth (Karlsruhe, Germany), Sigma-Aldrich (Steinheim, Germany), Fisher Scientific (Schwerte, Germany), Merck (Darmstadt, Germany), and OlbrichtArom (Leisnig, Germany). A derivative of the B. subtilis strain ATCC 6051 (American Type Culture Collection) was used for all chromosomal gene knockouts (Kabisch et al. 2013). Escherichia coli DH10B (Invitrogen, Darmstadt, Germany) [F-endA1 recA1 galE15 galK16 nupG rpsL ΔlacX74 Φ80lacZΔM15 araD139 Δ(ara,leu)7697 mcrA Δ(mrr-hsdRMS-mcrBC) λ-] was used as host strain for all cloning procedures. Sequences of primers used in cloning experiments of this study are summarized in Table S1. All strains created in this study are listed in Table 1. Plasmids constructed in this study are summarized in Table 2 and for the toolbox in Table S2 and S3.
All routine molecular biological techniques were carried out according to standard protocols (Sambrook and Russell 2001). Restriction enzymes and other DNA-modifying enzymes were used as specified by the suppliers (Roche, Mannheim, Germany; New England Biolabs, Frankfurt a. M., Germany). DNA sequencing was carried out by Eurofins Genomics (Ebersberg, Germany). Oligonucleotides were synthesized and provided by Life Technologies (Darmstadt, Germany).
Plasmid and strain constructions
Construction of B. subtilis LS8P-D
In order to construct an optimized B. subtilis expression strain, which is deficient in the eight main extracellular proteases and shows reduced autolysis as well as sporulation rates, the respective genes were deleted from the host genome using the Cre-loxP recombination technique as described by Kumpfmüller et al. (Kumpfmüller et al. 2013, 2016). For this purpose, B. subtilis strain BsJK49, carrying a Pxyl-cre, xylR, Pspac-comS, ZeoR cassette in the sacA locus for induction of the Cre recombinase, was used as the host strain for all further modifications. The vector backbone of pUC19 (Schweizer 1991) was used for all cloning procedures.
Deletion of the native lytC gene
For deletion of the native lytC gene of the B. subtilis host strain, the lox-SSS cassette was isolated from pJET-loxSSS (Kumpfmüller et al. 2013) and gel-purified using the Qiaquick Gel Extraction Kit (Qiagen, Hilden, Germany). The purified lox-SSS fragment was then ligated with the XbaI- and BamHI-restricted vector pUC19, and E. coli DH10B was transformed with the resulting recombinant plasmid, designated pUC19-loxSSS. The lytC-landing pads were amplified by PCR with oligonucleotides SO33/SO34 (lytC-front) and SO35/SO36 (lytC-back) using genomic DNA of B. subtilis as template. The lytC-front fragment was digested with SphI/XbaI and ligated with the SphI/XbaI-digested vector pUC19-lox-SSS. The recombinant plasmid was used for transformation of E. coli DH10B, yielding pUC19-lox-SSS-lytCF. Subsequently, the BamHI/KpnI digested and purified lytC-back fragment was ligated with pUC19-lox-SSS-lytCF. The recombinant plasmid was used for transformation of E. coli DH10B and sequence identity of all three DNA fragments was validated. The resulting plasmid was designated pUC19-lox-SSS-FB-lytC. Chromosomal integration of the lytC knockout vector and subsequent recombination of the lox sites was carried out as described before (Kumpfmüller et al. 2013). The resulting strain was designated BsJK49 ΔlytC (Table 1).
Deletion of the spoIIGA and bpr genes
The deletion of the chromosomal copies of the B. subtilis spoIIGA and bpr genes was combined in one step since both genes are located immediately adjacent to each other within the host genome. The respective DNA fragment, comprising the lox-SSS cassette and both landing pads, was amplified from pSC-B-bpr-spo-lox-SSS (Table 2) using oligonucleotides SO21 and SO22. The corresponding PCR product was digested, gel-purified, and integrated into the BamHI and XbaI sites of pUC19. Escherichia coli DH10B was transformed with the recombinant plasmid pUC19-lox-SSS-FB-bpr-spo and sequence identity was validated. After transformation of BsJK49 ΔlytC, strain BsJK49 ΔlytC, ΔPI was obtained.
Deletion of the nprB, mpr, aprE, nprE, vpr, epr, and wprA genes
For nprB, epr, vpr, aprE, and nprE, the knockout of the genome-encoded protease genes was carried out by integration of the respective gene-specific upstream and downstream sequences in the vector backbone of pUC19-lox-SSS. The front regions of the aforementioned protease genes were amplified with oligonucleotides SO38/SO39 (epr), SO42/SO43 (nprB), SO46/SO47 (nprE), SO50/SO51 (aprE), and SO56/SO57 (vpr). After restriction with KpnI/BamHI and subsequent PCR product purification, all upstream fragments were ligated with the KpnI- and BamHI-restricted plasmid pUC19-lox-SSS. Escherichia coli DH10B was transformed with the resulting intermediate plasmids carrying the lox-SSS cassette and the respective upstream region. Amplification of the downstream regions was carried out using oligonucleotides SO40/SO41 (epr), SO44/SO45 (nprB), SO48/SO49 (nprE), SO52/SO53 (aprE), and SO58/SO59 (vpr). The XbaI/SphI digested and purified DNA fragments were ligated with the corresponding intermediate plasmid. Escherichia coli DH10B was transformed with the resulting plasmids, designated pUC19-lox-SSS-FB-epr, pUC19-lox-SSS-FB-nprB, pUC19-lox-SSS-FB-nprE, pUC19-lox-SSS-FB-aprE, and pUC19-lox-SSS-FB-vpr. For mpr and wprA deletions, the appropriate DNA fragments comprising the lox-SSS cassette as well as the upstream and downstream regions of either mpr or wprA were amplified from pSC-B-mpr-lox-SSS (using oligonucleotides SO29/SO30) and pSC-B-wprA-lox-SSS (with oligonucleotides SO17/SO18), respectively. The purified and BamHI/XbaI-digested DNA fragments were then separately ligated with the XbaI- and BamHI-restricted vector pUC19. Escherichia coli DH10B was transformed with the resulting recombinant plasmids, designated pUC19-lox-SSS-FB-mpr and pUC19-lox-SSS-FB-wprA. Sequence identity of all pUC19-knockout vectors was validated, and transformation of BsJK49 ΔlytC, ΔPI was carried out successively. The final strain, B. subtilis LS8P-D, deficient in eight proteases and characterized by reduced autolysis and sporulation rates was obtained by replacing the Cre-ZeoR-cassette in the sacA locus of BsJK49 ΔlytC, ΔPVIII with the sequence of the spectinomycin selection marker. To this purpose, the integrative plasmid pJK256 (Zobel et al. 2015) was used. All gene deletions in the final B. subtilis LS8P-D strain as well as in all intermediate strains were confirmed by PCR in comparison to the unmodified parental strain BsJK49 and subsequent sequencing of the PCR products.
Deletion of secondary metabolite gene clusters
Further modification of the B. subtilis LS8P-D precursor strain BsJK49 ΔlytC, ΔPVIII was carried out by deletion of three big gene clusters for major secondary metabolite production. First the srfA cluster, responsible for the biosynthesis of surfactin was deleted using pJK191 (Kumpfmüller et al. 2016) (Table 2). This vector contained the lox-SSS cassette flanked by the homology regions for the srfA cluster. After transformation of BsJK49 ΔlytC, ΔPVIII, and subsequent induction of Cre recombinase, srfA deletion was confirmed by PCR and sequencing. The resulting strain was designated BsJK135. The subsequent deletions of the pksX gene cluster for bacillaene biosynthesis and the pps cluster for pliplastatin biosynthesis were carried out with pJK179 and pJK254 (Kumpfmüller et al. 2016) respectively. One after the other, the deletion vectors were used for transformation of BsJK135 (ΔsrfA) and BsJK136 (ΔsrfA, ΔpksX), yielding strain BsJK137 (ΔsrfA, ΔpksX, Δpps). All deletions were verified by PCR and sequencing. Additionally, to ensure cultivation in EnpressoB medium, the amylase gene in BsJK137 was deleted by transformation of the strain with plasmid pAMY-lox-SSS (Kumpfmüller et al. 2013), and the resulting strain was designated BsJK138. Finally, the Cre cassette was removed from the genome of BsJK138 using pJK256 as described above. This further evolved strain, BsJK139, was verified by PCR. The relevant genotypes of BsJK139 and all intermediate strains are listed in Table 1.
Construction of the plasmid library for Sox activity screening
In order to obtain a sox plasmid library with 173 different types of signal peptide (SP) DNA sequences in the required size of at least 2000 E. coli clones, the “B. subtilis Secretory Protein Expression System” (Takara/Clontech) in combination with the “In Fusion HD Cloning Plus Kit” (Takara/Clontech) was used according to the manufacturer’s instructions. To this end, the sox gene was amplified from pSox (Table 2) using oligonucleotides TB20 and TB21. After restriction with NdeI and XbaI, the purified PCR product and the pBE-S vector were ligated and E. coli DH10B was transformed with the recombinant plasmid pBE-S-sox. After validation of sequence identity, all different signal peptide sequences included in the provided SP library were integrated into the vector backbone of pBE-S-sox following the manufacturer’s instructions. In brief, the EagI- and MluI-digested vector was ligated with the 173 SP-containing DNA mixture using the “In Fusion Cloning” technology. Chemically competent E. coli Stellar cells (included in the kit) were transformed with 2 μL of the “In Fusion” reaction and selected on LB agar plates with ampicillin. All colony-forming units (cfu) were rinsed from the plate to isolate the sox plasmid library, which was subsequently integrated into the expression host B. subtilis LS8P-D ΔamyE by electroporation.
Cultivation conditions and media
Sox expression-prescreening experiments (Fig. S2) were carried out in 48-well plates (1-mL scale). Production strains that were identified in the screening experiments were then cultivated in 24-well deep-well plates for comparative sox expression (3.5-mL scale). For this purpose, EnpressoB medium was used for the expression studies with the host strain B. subtilis LS8P-D ΔamyE following the manufacturer’s instructions. Briefly, for each cultivation, an LB pre-culture was grown for 6–8 h at 250 rpm and 37 °C and was subsequently used to inoculate the main culture. The main culture was grown at 30 °C and 37 °C for 15–18 h. The booster solution was added in a 1:10 ratio and growth was continued for another 2 days. Cell harvesting was carried out 24 h and 48 h after addition of the booster solution. In case of all PacoA-regulated expression systems, the addition of the inducer acetoin (final concentration 1%) was necessary and occurred simultaneously with the addition of the booster solution. All cultivation and protein assay experiments were performed with at least three biological replicates.
Protein purification
For Sox activity screening, His-tag-based purification was realized using the HisSorb Plates (Qiagen) as recommended by the manufacturer. To this end, 200 μL of cell supernatant was bound to the plate surface, washed, and eluted in a final volume of 50 μL.
For determination of enzymatic activities derived from the cultivations in 3.5-mL and 1.5-L scale, ProCatch His Resin (Miltenyi Biotec) and Roti®garose-His/Ni Beads (Carl Roth) were used following the manufacturers’ instructions. In brief, 1 mL of the culture supernatant was incubated with 500 μL of His Resin for at least 20 min. After protein binding and washing procedures, the purified Sox protein was eluted from the His Resin with 200 μL elution buffer and was subsequently used for activity measurements.
Protein analysis
SDS-PAGE was carried out as described by Laemmli (Laemmli 1970). Four percent stacking gels and 15% or 18% separating gels were used depending on the size of the detected protein. Protein samples (20 μL) were mixed with 4 μL of 4 × sample buffer, denatured for 10 min at 95 °C, and loaded on the gel. SDS gel staining was carried out using Coomassie staining solution.
Protein quantification
If protein amounts are quantified from the culture supernatant, no dilution or concentration steps can impair the results. Thus, a realistic condition is depicted. For protein quantification, the gels were consistently loaded with all triplicate samples from the biological replicates, a molecular weight standard, and four dilutions of a BSA standard with known concentrations of 25, 50, 100, and 200 μg/mL. Based on the calibration curve derived from the BSA standard, the signal intensities of all protein samples were converted into protein amounts. To this end, the GelAnalyzer 2010 software was used. Based on the obtained data, the amounts of Sox and interleukin derived from all 12 expression strains were calculated and compared.
Results
The production of oxidative enzymes like the sulfhydryl oxidase Sox represents a challenge. These enzymes are able to produce reactive oxygen species as part of their catalyzed enzymatic reaction, which lead to damages of host cell components and concomitant low enzyme yields. Besides sophisticated and expensive small-scale cell-free production of these potential toxic proteins, secretion of the oxidative target enzymes to the medium was found to be one option to overcome part of the toxicity problem. Thus, Bacillus strains enabling extracellular protein accumulation are promising host candidates for the production of such oxidative enzymes for biotechnological applications. We therefore tested B. subtilis as a secretory expression host for the synthesis of the sulfhydryl oxidase Sox.
Development of a vector toolbox for B. subtilis
A successful secretory overexpression of foreign proteins, especially of eukaryotic origin, in bacterial expression systems cannot be coherently performed based on one standardized expression system alone. Due to the singularity of each individual heterologous protein, different host-vector systems and expression strategies have to be tested for optimal secretory overproduction. Therefore, we developed a vector toolbox for B. subtilis, which combines different strong secretion signals, vector backbones for the regulation of the gene copy number, two different inducible promoters, and a translational enhancer and a strong terminator sequence (Fig. 1). The construction of the plasmids in the toolbox is described in the Supplementary Online Material of this study. As backbone for this new host-vector system, we constructed the optimized B. subtilis expression strain LS8P-D, which is sporulation deficient and contains markerless gene deletions for eight main extracellular proteases (Table 1). After inactivation of the lytC gene, this strain shows reduced cell autolysis (Kabisch et al. 2013). Since potential bioactive compounds could be problematic in selected bioprocesses or in the application of overproduced enzymes, this strain was further modified by the deletion of three gene clusters responsible for the formation of the prominent secondary metabolites surfactin, bacillaene, and pliplastatin. The resulting strain B. subtilis BsJK139 was used in all experiments for the evaluation of the toolbox vectors constructed in this study.
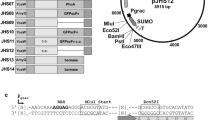
Schematic overview (a) of the replaceable elements of the B. subtilis toolbox and (b) the array of the toolbox elements including restriction sites. The multiple cloning site of all toolbox vectors was adjusted as shown in Figure S1. GOI, gene of interest
As a first step in the development of the toolbox, screening experiments were performed in order to determine suitable strong peptide secretion signals for efficient Sox export (see Supplementary Online Material). Highest yields of secreted Sox proteins were observed using the signal peptides of Csn, NucB, YweA, and YlqB (Fig. 2, Fig. S2-S3). The calculated protein amounts significantly exceeded those obtained with the previously used amyE signal peptide up to fivefold (Fig. 2). The secretion signals of csn and ylqB, which mediated the largest Sox quantities were thus used for the toolbox setup.
Sox amounts from SP screening. High amounts of Sox protein after 24 h (a) and 48 h (b) were calculated for the secretion signals of csn, nucB, yweA, and ylqB in comparison to significantly reduced amounts of Sox secreted by the amyE signal peptide. Dark blue bars represent Sox yields in μg/mL based on the quantification using a BSA standard (see Fig. S3b) while light blue bars illustrate the percentage distribution using the amyE-SP as reference (100%)
In addition, two further strong secretion signals deduced from the B. subtilis lipA and yncM genes were chosen for the toolbox based on literature data (Brockmeier et al. 2006). The toolbox enables modulation of the gene copy number by three vector backbones with high (pMSE3: 200–250 copies) (Silbersack et al. 2006), medium (pBE-S: 50 copies) (Leonhardt 1990), and low (pHB201: 5 copies) (Bron et al. 1987, 1998) copy numbers. Furthermore, the toolbox is based on the transient phase induced promoter (PaprE) of the B. subtilis subtilisin-encoding aprE gene (Veening et al. 2008) and the strictly regulated acetoin-inducible promoter (PacoA) of the B. subtilis acetoin dehydrogenase-encoding acoABCD operon (Silbersack et al. 2006). In addition, the downstream box (DB) of the B. subtilis cspB gene was included as a potential translational enhancer (Welsch et al. 2015). The strong transcriptional terminator structure derived from the B. subtilis bkd operon (Nickel et al. 2004) was chosen to prevent a potential transcriptional read through by the two strong promoters (Fig. 1a). Together with the possibility of intracellular protein production, this newly constructed toolbox consists of 60 vector variants (Table S2), each of which offers the possibility to easily exchange every single module by Gibson cloning or classical restriction and ligation (Fig. 1b).
Verification of the toolbox by Sox production
Using the sulfhydryl oxidase Sox (Lee et al. 2000) of S. cerevisiae and the human interleukin-1β (IL) (Garlanda et al. 2013) as model heterologous proteins, the suitability of the vector toolbox for B. subtilis was evaluated. To gain an overview about the influence of different toolbox modules on the yield of Sox, a set of 12 expression strains (see Table 3) for this target protein was analyzed under small-scale conditions in EnpressoB medium (Fig. 3a–c). Our signal peptide prescreening experiments indicated that most efficient Sox secretion was mediated by the csn signal peptide. Consequently, this specific SP was used for the construction of the individual Sox-producing strains based on the toolbox (Table 3, Table S2). The generated toolbox strains 1–4 carry the high-copy vector backbone pMSE3, strains 5–8 the medium-copy backbone pBE-S, and strains 9–12 the low-copy vector pHB201. The four variants of each backbone differ in the promoter sequences (PacoA for the numbers 1, 2, 5, 6, 9, 10, and PaprE for the remaining 3, 4, 7, 8, 11, 12 strains) and in the presence (uneven numbers) or absence (even numbers) of the DB enhancer sequence (see Table 3).
Verification of the toolbox strains S1–S12 (see also Table 3) for the overexpression of Sox. a–c Growth curves of B. subtilis JK138 S1–12 in comparison to the negative control carrying the “empty vector” without expression cassette. Cultivations were performed in biological triplicates; standard deviations are presented as error bars. d–f Quantitative analyses of the secretory overproduction of Sox. Average protein amounts (normalized by OD) for the Sox strains S1–S12 in the culture supernatant 24 h and 48 h after the boost in EnpressoB medium. Protein concentrations are given as average values from three independent experiments. Standard deviations are presented as error bars. For comparison of protein yields, the target protein was quantified. “Empty vector” strains without expression cassette served as negative controls
Sox quantification from the culture supernatants revealed pronounced differences in sox expression of the different vector setups, especially 24 h after the boost (Fig. 3d–f). For the pMSE3-carrying strains, highest Sox amounts (~ 12–15 μg/mL) were calculated for the PacoA-based expression strains (S1, S2), whereas significantly lower concentrations of approx. 5–7 μg/mL were calculated for the PaprE-based expression strains S3 and S4. The enhancer sequence in the strains S1 and S3 had a noticeable effect on protein production in comparison to their DB-free analogues S2 and S4. In contrast, within the pBE-S backbone, the absence of the enhancer sequence in S6 and S8 resulted in higher protein amounts compared to the analogous strains without DB (S5, S7) at t24h. In this background, highest secreted protein amounts were determined for S8 with 12.4 μg/mL. With average values of approximately 4 μg/mL, all low-copy vector-based expression strains S9-12 exhibited comparable but lower Sox yields. Forty-eight hours after the boost (Fig. 3d), the S1 and S2 strains still reached higher Sox concentrations compared to S3 and S4. However, slightly increased values were calculated for the strain variants without the enhancer sequence. The pBE-S-based strains S5-7 (Fig. 3e) showed similar protein amounts of about 10 μg/mL, whereas the Sox concentration for strain S8 without the enhancer sequence reached up to 17 μg/mL. The lowest protein amounts (approx. 5 μg/mL) were again determined for the S9-S12 strains which harbor the low-copy expression vectors (Fig. 3f). Thus, the highest sox expression was constantly reached by the high-copy vector with the acetoin-controlled PacoA promoter in strain S1 (Fig. 3d).
The toolbox was also evaluated by the overproduction of the human interleukin-1β (IL) (see Supplementary Online Material). The experimental setup for this second model protein was equal to the sox expression procedures. It is shown that the IL constructs gave similar results as the sox toolbox strains (Fig. S4). The highest IL production was reached with the IL2 toolbox setup containing the high-copy vector and the acetoin-controlled PacoA promoter.
Determination of Sox activity
In order to determine a functional expression of the sulfhydryl oxidase we established a suitable procedure for Sox activity analyses. Culture supernatants of the model strain B. subtilis WB600 pSox were concentrated by lyophilization and subsequent resuspension of the extracellular protein extracts in water. The pH of the obtained protein solution was adjusted to 8.0 and used for His-tag-based affinity chromatography. Sox purification from the supernatant was monitored by SDS-PAGE (Fig. 4a). Purified Sox proteins were used to determine temperature and pH profiles of this heterologously produced enzyme (Fig. 4b). Maximum Sox activity was determined at 30 °C and at alkaline pH ranging from pH 9 to 11.
Activity of the heterologously produced sulfhydryl oxidase Sox. a Purification of Sox from the supernatant of B. subtilis WB600 pSox cultures; SN, supernatant; FT, flow through, W1–W2: wash fractions, E1–E4: elution fractions. b Temperature and pH profiles of purified Sox. The influence of temperature (light blue dots) and pH (dark blue dots) on Sox activity was determined using a miniaturized Sox assay in a semi-automated system applying a Tecan Freedom EVO® screening robot
Fermentation strategies for Sox production
Essential fermentation parameters for heterologous sox expression in B. subtilis were identified and stepwise optimized. The small-scale cultivations indicated that the PacoA promoter was most suitable for Sox (Fig. 3) as well as IL (Fig. S4) production. Therefore, we used this acetoin-inducible promoter system to develop a fermentation strategy for the Sox target protein. We investigated the influence of 30 °C vs. 37 °C as growth temperature on enzyme yield with this promoter system (Fig. S5). Although similar cell densities were measured after 12 h of cultivation, a faster growth in combination with an earlier start of Sox production was observed at 37 °C. We therefore performed all following fermentations at 37 °C. Furthermore, we tested the influence of different glucose concentrations on growth and productivity of B. subtilis WB600 pSox (Fig. S6). Our analyses revealed that both investigated glucose concentrations enabled similar Sox activities and final optical densities.
We therefore used these temperature and glucose parameters to develop a fed-batch fermentation strategy, which was compared to the established batch process (Fig. 5a). The batch fermentation was performed again with NBMM containing threefold concentrated complex components yeast extract and peptone (45 and 9.6 g/L, respectively), 0.5% (w/v) acetoin and 10 g/L glucose. The fed-batch fermentation was carried out with a starting batch containing NBMM medium with 15 g/L yeast extract, 3.2 g/L peptone, 0.5% (w/v) acetoin, and 10 g/L glucose. Feed I was started at 4-h fermentation time with 7.5 g/h yeast extract, 1.6 g/h peptone, and 4.0 g/h glucose. Feed II was started at 7-h fermentation time with 7.5 g/h yeast extract and 1.6 g/h peptone. Growth curves and Sox activities were determined as before and showed that the consecutive feeding strategy applied in the fed-batch fermentation yielded maximum optical densities of 33.0 after 12 h compared to 29.4 after 16 h in the batch fermentation and seemed to result in higher autolysis rates. A significant increase in Sox activity in the fermentation supernatant of 76.1 U/L after 24 h was determined when applying a fed-batch rather than a batch fermentation strategy, which yielded 60.8 U/ L after 24 h (Fig. 5a). This was confirmed by SDS-PAGE analyses which revealed increased amounts of Sox protein during the fed-batch fermentation in the total extracellular protein extract. Thus, the fed-batch strategy enabled higher Sox activities and levels compared to the batch process (Fig. 5).
Development of a fermentation strategy for Sox overproduction. a Comparison of batch (light blue) and fed-batch (dark blue) fermentation strategies for Sox production in B. subtilis WB600 pSox. The cells were grown in a parallel fermenter system at 1.5 L scale at 37 °C either in batch or in fed-batch mode. Cell growth was monitored by the OD at 600 nm (triangles). Sox activity was determined in triplicates using purified enzyme (bars). b SDS-PAGE analysis of Sox accumulation in untreated culture supernatants of the batch and fed-batch fermentations
The cultivation experiments with the protease-deficient B. subtilis strain WB600 revealed an autolysis of the cells throughout the fermentation, especially at 37 °C and during the stationary phase. For this reason, Sox production of B. subtilis WB600 pSox was compared with the novel tailor-made expression strain LS8P-D pSox, which possesses not only clean gene deletions of the eight major extracellular proteases and three active secondary metabolite clusters but which is also deficient in the lytC gene (Fig. 6a). In these experiments, the NBMM medium was supplemented with twofold concentrated yeast extract and peptone (30 and 6.4 g/L, respectively) and 2 g/L glucose. Since additional tests in small-scale showed that reduction of the inductor acetoin was possible without decrease in productivity (data not shown), only 0.05% (w/v) acetoin was used to induce Sox production. This medium composition resulted in a slightly higher final optical density of B. subtilis LS8P-D pSox compared to the WB600 pSox strain (Fig. 6a). This higher final optical density and the more stable stationary phase of the B. subtilis LS8P-D pSox cultivations indicated reduced autolysis of this novel strain. Furthermore, compared to the WB600 strain, the engineered LS8P-D expression strain revealed similar Sox activities in the extracellular protein supernatant (Fig. 6a).
a Comparison of growth and Sox production of the protease-deficient B. subtilis strains WB600 (light blue) and LS8P-D (dark blue). The cells were grown in batch fermentations at 37 °C (triangles). Sox activity was determined in triplicates using purified enzyme after 24 h (bars). b Verification of the best performing PacoA-dependent S1 toolbox system in B. subtilis JK139 for the overexpression of the sulfhydryl oxidase Sox. The growth curve reflects three individual 1.5-L fed-batch fermentation experiments A–C. Shown are average protein amounts (normalized by OD) from the culture supernatant of the S1 expression strains from three independent experiments. Standard deviations are presented as error bars
We finally used this B. subtilis LS8P-D mutant background in form of the novel B. subtilis mutant strain BsJK139 and the established large-scale fed-batch fermentation strategy to test the overproduction of the best performing Sox and IL toolbox setups. In both expression strains, the high-copy vector pMSE3 was used, and in the case of the Sox-producing setup, S1 the DB enhancer sequence was included. Each of the 1.5-L fed-batch cultivations for either Sox or IL production was carried out in triplicates. Samples for protein analyses were taken after 12, 24, 36, and 48 h (Fig. 6b, Fig. S7). Expression of the sox gene was induced by initial addition of 0.5% (w/v) acetoin. The calculated Sox amounts in the supernatant of all three fermentations showed a continuous increase in protein concentrations from 1.8 (t12h) to 6.5 μg/mL (t48h) over the whole fermentation time (Fig. 6b). Compared to the small-scale cultivations, however, slightly lower amounts of the Sox target protein were produced. We tested the same fed-batch fermentation strategy for the best performing IL producing toolbox setup IL2 in the novel B. subtilis BsJK139 expression strain by using the newly established large-scale fed-batch strategy (Fig. S7). A similar expression pattern of the interleukin was determined in these cultivation experiments. However, in contrast to Sox production, comparable IL protein amounts were calculated in the small- and large-scale fed-batch cultivations.
Discussion
The aim of this study was to develop a versatile expression platform for the secretory production of heterologous proteins in B. subtilis. The different copy numbers of the three vector backbones of the established toolbox enable a sensitive fine-tuning of target gene expression by affecting the amounts of transcripts which, in turn, determine the rate of target protein synthesis. However, high-copy numbers of plasmids can also negatively influence cell physiology due to the increased metabolic demands by plasmid DNA and mRNA synthesis as well as protein translation of encoded proteins. Such a cellular metabolic burden may lead to reduced capacities for target protein production or plasmid instabilities. Thus, a reasonable balance between plasmid copy number, expression strength of the chosen promoter, and recombinant protein synthesis is required for efficient protein production processes. We observed that the copy number of dependent expression of the two model genes of this study resulted in different expression rates by the toolbox vectors. Whereas the Sox protein reached highest secreted protein levels with the multi-copy vector backbone of pMSE3 or medium-copy plasmid pBE-S, the interleukin revealed rather weaker protein amounts with these high-copy variants but a slightly higher protein level with the low-copy vector pHB201.
Our toolbox analyses also indicated that the acoA promoter leads in most cases to a higher expression level of the two target genes compared to the aprE promoter. This observed trend for Sox overproduction might be partially due to the strict control of the acoA promoter which needs the inducer acetoin, but which is also only strongly active if the preferred carbon source glucose is exhausted (Silbersack et al. 2006). However, the aprE promoter that is controlled by DegU and induced during nutrient limitation (Veening et al. 2008) might also be competitive if an optimized fermentation strategy is used. Especially for the production of potential toxic enzymes, the PacoA variants might be advantageous. A premature induction of the acoA promoter is repressed in the presence of glucose during exponential growth (Silbersack et al. 2006) and higher cell densities of the expression host prior to induction could be beneficial to ensure an efficient synthesis of critical proteins (Müller et al. 1996).
The in this study established toolbox (Table S2) with three different expression vector backbones, two promoters, four secretion signals, and the presence or absence of the translational enhancer sequence comprises 60 plasmids. Each module of this plasmid system can easily be exchanged by standard molecular cloning or assembly methods (Fig. 1b). This toolbox allows individual adjustments of heterologous protein expression and secretion, which was successfully evaluated by means of the two model proteins, sulfhydryl oxidase from S. cerevisiae and human interleukin-1β in this study.
Finally, the application of the novel toolbox is supported by the prototrophic protease and sporulation-deficient strain BsJK139, which allows for improved downstream processing by reduced secondary metabolism and autolysis, and prevents premature proteolysis of target proteins. Although the obtained Sox yield is relatively low compared to other target proteins overproduced in Bacilli (Lakowitz et al. 2018), the combination of this engineered B. subtilis strain background with the tightly regulated acoA promoter and the established glucose- and acetoin-controlled fed-batch fermentation strategy enabled a significant overproduction of this potentially toxic sulfhydryl oxidase in an active form. In summary, the proposed expression toolbox in combination with the optimized host cell backbone ensures efficient recombinant production of proteins that are difficult-to-express in B. subtilis.
Data availability
The strains and plasmids constructed in this study as well as the data that support the findings of this study are available from the corresponding author upon request.
Code availability
Not applicable.
References
Aguilar Suárez R, Stülke J, van Dijl JM (2019) Less is more: toward a genome-reduced Bacillus cell factory for “difficult proteins.” ACS Synth Biol 8:99–108. https://doi.org/10.1021/acssynbio.8b00342
Brockmeier U, Caspers M, Freudl R, Jockwer A, Noll T, Eggert T (2006) Systematic screening of all signal peptides from Bacillus subtilis: a powerful strategy in optimizing heterologous protein secretion in Gram-positive bacteria. J Mol Biol 362:393–402. https://doi.org/10.1016/j.jmb.2006.07.034
Bron S, Bolhuis A, Tjalsma H, Holsappel S, Venema G, van Dijl JM (1998) Protein secretion and possible roles for multiple signal peptidases for precursor processing in Bacilli. J Biotechnol 64:3–13. https://doi.org/10.1016/S0168-1656(98)00099-6
Bron S, Bosma P, van Belkum M, Luxen E (1987) Stability function in the Bacillus subtilis plasmid pTA1060. Plasmid 18:8–15. https://doi.org/10.1016/0147-619X(87)90073-4
Emond E, Lavallée R, Drolet G, Moineau S, LaPointe G (2001) Molecular characterization of a theta replication plasmid and its use for development of a two-component food-grade cloning system for Lactococcus lactis. Appl Environ Microbiol 67:1700–1709. https://doi.org/10.1128/AEM.67.4.1700-1709.2001
Faccio G, Nivala O, Kruus K, Buchert J, Saloheimo M (2011) Sulfhydryl oxidases: sources, properties, production and applications. Appl Microbiol Biotechnol 91:957–966. https://doi.org/10.1007/s00253-011-3440-y
Fan X, Wu H, Jia Z, Li G, Li Q, Chen N, Xie X (2018) Metabolic engineering of Bacillus subtilis for the co-production of uridine and acetoin. Appl Microbiol Biotechnol 102:8753–8762. https://doi.org/10.1007/s00253-018-9316-7
Garlanda C, Dinarello CA, Mantovani A (2013) The interleukin-1 family: back to the future. Immunity 39:1003–1018. https://doi.org/10.1016/j.immuni.2013.11.010
Harwood CR, Cranenburgh R (2008) Bacillus protein secretion: an unfolding story. Trends Microbiol 16:73–79. https://doi.org/10.1016/j.tim.2007.12.001
Kabisch J, Thürmer A, Hübel T, Popper L, Daniel R, Schweder T (2013) Characterization and optimization of Bacillus subtilis ATCC 6051 as an expression host. J Biotechnol 163:97–104. https://doi.org/10.1016/j.jbiotec.2012.06.034
Kumpfmüller J, Kabisch J, Schweder T (2013) An optimized technique for rapid genome modifications of Bacillus subtilis. J Microbiol Methods 95:350–352. https://doi.org/10.1016/j.mimet.2013.10.003
Kumpfmüller J, Methling K, Fang L, Pfeifer BA, Lalk M, Schweder T (2016) Production of the polyketide 6-deoxyerythronolide B in the heterologous host Bacillus subtilis. Appl Microbiol Biotechnol 100:1209–1220. https://doi.org/10.1007/s00253-015-6990-6
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685. https://doi.org/10.1038/227680a0
Lakowitz A, Godard T, Biedendieck R, Krull R (2018) Mini review: recombinant production of tailored bio-pharmaceuticals in different Bacillus strains and future perspectives. European journal of pharmaceutics and biopharmaceutics : official journal of Arbeitsgemeinschaft fur Pharmazeutische Verfahrenstechnik e.V 126:27–39. https://doi.org/10.1016/j.ejpb.2017.06.008
Lee J-E, Hofhaus G, Lisowsky T (2000) Erv1p from Saccharomyces cerevisiae is a FAD-linked sulfhydryl oxidase. FEBS Lett 477:62–66. https://doi.org/10.1016/S0014-5793(00)01767-1
Leonhardt H (1990) Identification of a low-copy-number mutation within the pUB110 replicon and its effect on plasmid stability in Bacillus subtilis. Gene 94:121–124. https://doi.org/10.1016/0378-1119(90)90477-9
Liu S, Endo K, Ara K, Ozaki K, Ogasawara N (2008) Introduction of marker-free deletions in Bacillus subtilis using the AraR repressor and the ara promoter. Microbiology (reading) 154:2562–2570. https://doi.org/10.1099/mic.0.2008/016881-0
Liu X, Wang H, Wang B, Pan L (2018a) Efficient production of extracellular pullulanase in Bacillus subtilis ATCC6051 using the host strain construction and promoter optimization expression system. Microb Cell Fact 17:163. https://doi.org/10.1186/s12934-018-1011-y
Liu X, Wang H, Wang B, Pan L (2018b) High-level extracellular protein expression in Bacillus subtilis by optimizing strong promoters based on the transcriptome of Bacillus subtilis and Bacillus megaterium. Protein Expr Purif 151:72–77. https://doi.org/10.1016/j.pep.2018.06.006
Müller J, van Dijl JM, Venema G, Bron S (1996) Cloning of heterologous genes specifying detrimental proteins on pUC-derived plasmids in Escherichia coli. Mol Gen Genet 252:207–211. https://doi.org/10.1007/BF02173221
Nickel M, Homuth G, Böhnisch C, Mäder U, Schweder T (2004) Cold induction of the Bacillus subtilis bkd operon is mediated by increased mRNA stability. Mol Genet Genomics 272:98–107. https://doi.org/10.1007/s00438-004-1038-0
Popp PF, Dotzler M, Radeck J, Bartels J, Mascher T (2017) The Bacillus BioBrick Box 2.0: expanding the genetic toolbox for the standardized work with Bacillus subtilis. Sci Rep 7:15058. https://doi.org/10.1038/s41598-017-15107-z
Radeck J, Kraft K, Bartels J, Cikovic T, Dürr F, Emenegger J, Kelterborn S, Sauer C, Fritz G, Gebhard S, Mascher T (2013) The Bacillus BioBrick Box: generation and evaluation of essential genetic building blocks for standardized work with Bacillus subtilis. J Biol Eng 7:29. https://doi.org/10.1186/1754-1611-7-29
Rosales-Mendoza S, Angulo C, Meza B (2016) Food-grade organisms as vaccine biofactories and oral delivery vehicles. Trends Biotechnol 34:124–136. https://doi.org/10.1016/j.tibtech.2015.11.007
Sambrook J, Russell DW (2001) Molecular cloning. A laboratory manual, 3rd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor (New York)
Schallmey M, Singh A, Ward OP (2004) Developments in the use of Bacillus species for industrial production. Can J Microbiol 50:1–17. https://doi.org/10.1139/w03-076
Schweizer HP (1991) Escherichia-Pseudomonas shuttle vectors derived from pUC18/19. Gene 97:109–112. https://doi.org/10.1016/0378-1119(91)90016-5
Silbersack J, Jürgen B, Hecker M, Schneidinger B, Schmuck R, Schweder T (2006) An acetoin-regulated expression system of Bacillus subtilis. Appl Microbiol Biotechnol 73:895–903. https://doi.org/10.1007/s00253-006-0549-5
Taguchi S, Ooi T, Mizuno K, Matsusaki H (2015) Advances and needs for endotoxin-free production strains. Appl Microbiol Biotechnol 99:9349–9360. https://doi.org/10.1007/s00253-015-6947-9
Toymentseva AA, Schrecke K, Sharipova MR, Mascher T (2012) The LIKE system, a novel protein expression toolbox for Bacillus subtilis based on the liaI promoter. Microb Cell Fact 11:143. https://doi.org/10.1186/1475-2859-11-143
van Dijl JM, Hecker M (2013) Bacillus subtilis: from soil bacterium to super-secreting cell factory. Microb Cell Fact 12:3. https://doi.org/10.1186/1475-2859-12-3
van Tilburg AY, van Heel AJ, Stülke J, de Kok NAW, Rueff A-S, Kuipers OP (2020) MiniBacillus PG10 as a convenient and effective production host for lantibiotics. ACS Synth Biol 9:1833–1842. https://doi.org/10.1021/acssynbio.0c00194
Veening J-W, Igoshin OA, Eijlander RT, Nijland R, Hamoen LW, Kuipers OP (2008) Transient heterogeneity in extracellular protease production by Bacillus subtilis. Mol Syst Biol 4:184. https://doi.org/10.1038/msb.2008.18
Welsch N, Homuth G, Schweder T (2015) Stepwise optimization of a low-temperature Bacillus subtilis expression system for “difficult to express” proteins. Appl Microbiol Biotechnol 99:6363–6376. https://doi.org/10.1007/s00253-015-6552-y
Wu S-C, Yeung JC, Duan Y, Ye R, Szarka SJ, Habibi HR, Wong S-L (2002) Functional production and characterization of a fibrin-specific single-chain antibody fragment from Bacillus subtilis: effects of molecular chaperones and a wall-bound protease on antibody fragment production. Appl Environ Microbiol 68:3261–3269. https://doi.org/10.1128/aem.68.7.3261-3269.2002
Wu XC, Lee W, Tran L, Wong SL (1991) Engineering a Bacillus subtilis expression-secretion system with a strain deficient in six extracellular proteases. J Bacteriol 173:4952–4958. https://doi.org/10.1128/jb.173.16.4952-4958.1991
Yang S, Kang Z, Cao W, Du G, Chen J (2016) Construction of a novel, stable, food-grade expression system by engineering the endogenous toxin-antitoxin system in Bacillus subtilis. J Biotechnol 219:40–47. https://doi.org/10.1016/j.jbiotec.2015.12.029
Yu H, Yan X, Shen W, Shen Y, Zhang J, Li S (2010) Efficient and precise construction of markerless manipulations in the Bacillus subtilis genome. J Microbiol Biotechnol 20:45–53. https://doi.org/10.4014/jmb.0904.04051
Zemella A, Thoring L, Hoffmeister C, Kubick S (2015) Cell-free protein synthesis: pros and cons of prokaryotic and eukaryotic systems. ChemBioChem 16:2420–2431. https://doi.org/10.1002/cbic.201500340
Zhang K, Su L, Wu J (2018) Enhanced extracellular pullulanase production in Bacillus subtilis using protease-deficient strains and optimal feeding. Appl Microbiol Biotechnol 102:5089–5103. https://doi.org/10.1007/s00253-018-8965-x
Zhao L, Ye B, Zhang Q, Cheng D, Zhou C, Cheng S, Yan X (2019) Construction of second generation protease-deficient hosts of Bacillus subtilis for secretion of foreign proteins. Biotechnol Bioeng 116:2052–2060. https://doi.org/10.1002/bit.26992
Zobel S, Kumpfmüller J, Süssmuth RD, Schweder T (2015) Bacillus subtilis as heterologous host for the secretory production of the non-ribosomal cyclodepsipeptide enniatin. Appl Microbiol Biotechnol 99:681–691. https://doi.org/10.1007/s00253-014-6199-0
Acknowledgements
The authors would like to thank all members of the Institute of Technical Microbiology at TUHH and the Institute of Pharmacy at University of Greifswald, for valuable input, discussions, and support during the course of the project. We thank especially Stephanie Markert for critical reading of the manuscript.
Funding
Open Access funding enabled and organized by Projekt DEAL. This study was financially supported by the German Federal Ministry of Education and Research (BMBF; reference 0315400A/B) in the frame of the BIOCATALYSIS2021 network and by the Ministry of Economy, Labour and Tourism of Mecklenburg Vorpommern and the European Fund for Regional Development (V-630-S-177–2013/213; V-630-F-177–2013/212; V630-VB305-2013/211).
Author information
Authors and Affiliations
Contributions
A. K. and N. W. performed most of the experiments and wrote the manuscript. T. S. initiated and coordinated this study and revised the paper. A. D. and N. W. carried out all experiments for signal peptide screening. J. K. constructed the engineered B. subtilis LS8P-D strain. J. F. K. was responsible for the construction of the pSox vector. H. P. and S. B. have carried out fermentation studies and Sox activity tests. H. G. M. and H. P. performed small- and large-scale cultivations. R. W. and H. B. were involved in data analyzes. L. P. initiated and supported the Sox study. H. T. initiated and supported the IL study. G. A. was involved in project coordination. All authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Krüger, A., Welsch, N., Dürwald, A. et al. A host-vector toolbox for improved secretory protein overproduction in Bacillus subtilis. Appl Microbiol Biotechnol 106, 5137–5151 (2022). https://doi.org/10.1007/s00253-022-12062-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-022-12062-2