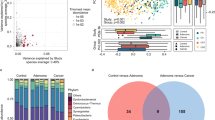
Abstract
Gut microbiota is a primary driver of inflammation in the colon and is linked to early colorectal cancer (CRC) development. Thus, a novel and noninvasive microbiome-based model could promote screening in patients at average risk for CRC. Nevertheless, the relevance and effectiveness of microbial biomarkers for noninvasive CRC screening remains unclear, and researchers lack the data to distinguish CRC-related gut microbiome biomarkers from those of other common gastrointestinal (GI) diseases. Microbiome-based classification distinguishes patients with CRC from normal participants and excludes other CRC-relevant diseases (e.g., GI bleed, adenoma, bowel diseases, and postoperative). The area under the receiver operator characteristic curve (AUC) was 92.2%. Known associations with oral pathogenic features, benefits-generated features, and functional features of CRC were confirmed using the model. Our optimised prediction model was established using large-scale experimental population-based data and other sequence-based faecal microbial community data. This model can be used to identify the high-risk groups and has the potential to become a novel screening method for CRC biomarkers because of its low false-positive rate (FPR) and good stability.
Key points
• A total of 5744 CRC and non-CRC large-scale faecal samples were sequenced, and a model was constructed for CRC discrimination on the basis of the relative abundance of taxonomic and functional features.
• This model could identify high-risk groups and become a novel screening method for CRC biomarkers because of its low FPR and good stability.
• The association relationship of oral pathogenic features, benefits-generated features, and functional features in CRC was confirmed by the study.






Similar content being viewed by others
Data availability
All sequencing datasets in this work are available for download at https://www.ncbi.nlm.nih.gov/bioproject/PRJNA396815.
References
Basch CH, Basch CE, Zybert P, Wolf RL (2016) Fear as a barrier to asymptomatic colonoscopy screening in an urban minority population with health insurance. J Commun Health 41(4):1–7. https://doi.org/10.1007/s10900-016-0159-9
Baxter NT, Ruffin MTT, Rogers MA, Schloss PD (2016) Microbiota-based model improves the sensitivity of fecal immunochemical test for detecting colonic lesions. Genome Med 8(1):37. https://doi.org/10.1186/s13073-016-0290-3
Candela M, Maccaferri S, Turroni S, Carnevali P, Brigidi P (2010) Functional intestinal microbiome, new frontiers in prebiotic design. Int J Food Microbiol 140(2–3):93–101. https://doi.org/10.1016/j.ijfoodmicro.2010.04.017
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7(5):335–336. https://doi.org/10.1038/nmeth.f.303
Caporaso JG, Lauber CL, Costello EK, Berg-Lyons D, Gonzalez A, Stombaugh J, Knights D, Gajer P, Ravel J, Fierer N, Gordon JI, Knight R (2011) Moving pictures of the human microbiome. Genome Biol 12(5):R50. https://doi.org/10.1186/gb-2011-12-5-r50
Carethers JM, Jung BH (2015) Genetics and genetic biomarkers in sporadic colorectal cancer. Gastroenterology 149(5):1177-e11901173
Chang WC, Gorecki P, Eulenstein O (2013) Exact solutions for species tree inference from discordant gene trees. J Bioinform Comput Biol 11(5):1342005. https://doi.org/10.1142/S0219720013420055
Chen C, Chen L, Lin L, Jin D, Du Y, Lyu J (2021) Research progress on gut microbiota in patients with gastric cancer, esophageal cancer, and small intestine cancer. Appl Microbiol Biotechnol 105(11):4415–4425. https://doi.org/10.1007/s00253-021-11358-z
Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ (2015) He J (2016) Cancer statistics in China. CA Cancer J Clin 66(2):115–132. https://doi.org/10.3322/caac.21338
Cho SY, Kim SH, Kang SH, Lee KJ, Choi D, Kang S, Park SJ, Kim T, Yoon CH, Youn TJ, Chae IH (2021) Pre-existing and machine learning-based models for cardiovascular risk prediction. Sci Rep 11(1):8886. https://doi.org/10.1038/s41598-021-88257-w
Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R (2009) Bacterial community variation in human body habitats across space and time. Science 326(5960):1694–1697. https://doi.org/10.1126/science.1177486
David LA, Materna AC, Friedman J, Campos-Baptista MI, Blackburn MC, Perrotta A, Erdman SE, Alm EJ (2014) Host lifestyle affects human microbiota on daily timescales. Genome Biol 15(7):R89. https://doi.org/10.1186/gb-2014-15-7-r89
Debelius JW, Vazquez-Baeza Y, McDonald D, Xu Z, Wolfe E, Knight R (2016) Turning participatory microbiome research into usable data: lessons from the American gut project. J Microbiol Biol Educ 17(1):46–50. https://doi.org/10.1128/jmbe.v17i1.1034
Ding M, Lang Y, Shu H, Shao J, Cui L (2021) Microbiota-gut-brain axis and epilepsy: a review on mechanisms and potential therapeutics. Front Immunol 12:742449. https://doi.org/10.3389/fimmu.2021.742449
Ding T, Schloss PD (2014) Dynamics and associations of microbial community types across the human body. Nature 509(7500):357–360. https://doi.org/10.1038/nature13178
Du Y, Wang H, Chen L, Fang Q, Zhang B, Jiang L, Wu Z, Yang Y, Zhou Y, Chen B, Lyu J, Wang Z (2021) Non-RBM mutations impaired SARS-COV-2 spike protein regulated to the ACE2 receptor based on molecular dynamic simulation. Front Mol Biosci 8:614443. https://doi.org/10.3389/fmolb.2021.614443
Faith JJ, Guruge JL, Charbonneau M, Subramanian S, Seedorf H, Goodman AL, Clemente JC, Knight R, Heath AC, Leibel RL, Rosenbaum M, Gordon JI (2013) The long-term stability of the human gut microbiota. Science 341(6141):1237439. https://doi.org/10.1126/science.1237439
Fushiki T (2011) Estimation of prediction error by using K -fold cross-validation. Stat Comput 21(2):137–146. https://doi.org/10.1007/s11222-009-9153-8
Giorgi Rossi P, Carretta E, Mangone L, Baracco S, Serraino D, Zorzi M, Colorectal Cancer Screening IWG (2018) Incidence of interval cancers in faecal immunochemical test colorectal screening programmes in Italy. J Med Screen 25(1):32–39. https://doi.org/10.1177/0969141316686391
Goodrich JK, Davenport ER, Beaumont M, Jackson MA, Knight R, Ober C, Spector TD, Bell JT, Clark AG, Ley RE (2016) Genetic determinants of the gut microbiome in UK twins. Cell Host Microbe 19(5):731–743. https://doi.org/10.1016/j.chom.2016.04.017
Goss PE, Strasser-Weippl K, Lee-Bychkovsky BL, Fan L, Li J, Chavarri-Guerra Y, Liedke PE, Pramesh CS, Badovinac-Crnjevic T, Sheikine Y, Chen Z, Qiao YL, Shao Z, Wu YL, Fan D, Chow LW, Wang J, Zhang Q, Yu S, Shen G, He J, Purushotham A, Sullivan R, Badwe R, Banavali SD, Nair R, Kumar L, Parikh P, Subramanian S, Chaturvedi P, Iyer S, Shastri SS, Digumarti R, Soto-Perez-de-Celis E, Adilbay D, Semiglazov V, Orlov S, Kaidarova D, Tsimafeyeu I, Tatishchev S, Danishevskiy KD, Hurlbert M, Vail C, St Louis J, Chan A (2014) Challenges to effective cancer control in China, India, and Russia. Lancet Oncol 15(5):489–538. https://doi.org/10.1016/S1470-2045(14)70029-4
Hamady M, Knight R (2009) Microbial community profiling for human microbiome projects: tools, techniques, and challenges. Genome Res 19(7):1141–1152. https://doi.org/10.1101/gr.085464.108
Hattori M, Taylor TD (2009) The human intestinal microbiome: a new frontier of human biology. DNA Res 16(1):1–12. https://doi.org/10.1093/dnares/dsn033
Human Microbiome Project C (2012) Structure, function and diversity of the healthy human microbiome. Nature 486(7402):207–214. https://doi.org/10.1038/nature11234
Imperiale TF, Ransohoff DF, Itzkowitz SH, Levin TR, Lavin P, Lidgard GP, Ahlquist DA, Berger BM (2014) Multitarget stool DNA testing for colorectal-cancer screening. N Engl J Med 370(14):1287–1297. https://doi.org/10.1056/NEJMoa1311194
Irrazabal T, Belcheva A, Girardin SE, Martin A, Philpott DJ (2014) The multifaceted role of the intestinal microbiota in colon cancer. Mol Cell 54(2):309–320. https://doi.org/10.1016/j.molcel.2014.03.039
Jess T, Horvath-Puho E, Fallingborg J, Rasmussen HH, Jacobsen BA (2013) Cancer risk in inflammatory bowel disease according to patient phenotype and treatment: a Danish population-based cohort study. Am J Gastroenterol 108(12):1869–1876. https://doi.org/10.1038/ajg.2013.249
Kalderstam J, Eden P, Ohlsson M (2015) Finding risk groups by optimizing artificial neural networks on the area under the survival curve using genetic algorithms. PLoS ONE 10(9):e0137597. https://doi.org/10.1371/journal.pone.0137597
Kuipers EJ, Rosch T, Bretthauer M (2013) Colorectal cancer screening–optimizing current strategies and new directions. Nat Rev Clin Oncol 10(3):130–142. https://doi.org/10.1038/nrclinonc.2013.12
Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Vega Thurber RL, Knight R, Beiko RG, Huttenhower C (2013) Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 31(9):814–821. https://doi.org/10.1038/nbt.2676
Levin B, Lieberman DA, McFarland B, Andrews KS, Brooks D, Bond J, Dash C, Giardiello FM, Glick S, Johnson D, Johnson CD, Levin TR, Pickhardt PJ, Rex DK, Smith RA, Thorson A, Winawer SJ, American Cancer Society Colorectal Cancer Advisory G, Force USM-ST, American College of Radiology Colon Cancer C (2008) Screening and surveillance for the early detection of colorectal cancer and adenomatous polyps, 2008: a joint guideline from the American cancer society, the US multi-society task force on colorectal cancer, and the American college of radiology. Gastroenterology 134(5):1570–1595. https://doi.org/10.1053/j.gastro.2008.02.002
Li JG, Wang CD, Tang ZH, Guo YQ, Zheng TC, Li YZ, You ZQ (2017) The gut bacterial community composition of wild Cervusalbirostris (white-lipped deer) detected by the 16S ribosomal RNA gene sequencing. Curr Microbiol 74(9):1100–1107. https://doi.org/10.1007/s00284-017-1288-9
Lozupone CA, Stombaugh J, Gonzalez A, Ackermann G, Wendel D, Vazquez-Baeza Y, Jansson JK, Gordon JI, Knight R (2013) Meta-analyses of studies of the human microbiota. Genome Res 23(10):1704–1714. https://doi.org/10.1101/gr.151803.112
Mao DP, Zhou Q, Chen CY, Quan ZX (2012) Coverage evaluation of universal bacterial primers using the metagenomic datasets. BMC Microbiol 12:66. https://doi.org/10.1186/1471-2180-12-66
Martin J, Halm EA, Tiro JA, Merchant Z, Balasubramanian BA, McCallister K, Sanders JM, Ahn C, Bishop WP, Singal AG (2017) Reasons for lack of diagnostic colonoscopy after positive result on fecal immunochemical test in a safety-net health system. Am J Med 130(1):93 e91-93 e97. https://doi.org/10.1016/j.amjmed.2016.07.028
Mazmanian SK, Round JL, Kasper DL (2008) A microbial symbiosis factor prevents intestinal inflammatory disease. Nature 453(7195):620–625. https://doi.org/10.1038/nature07008
Miller PJ, McArtor DB, Lubke GH (2017) A gradient boosting machine for hierarchically clustered data. Multivariate Behav Res 52(1):117. https://doi.org/10.1080/00273171.2016.1265433
Pang H, Lin A, Holford M, Enerson BE, Lu B, Lawton MP, Floyd E, Zhao H (2006) Pathway analysis using random forests classification and regression. Bioinformatics 22(16):2028–2036. https://doi.org/10.1093/bioinformatics/btl344
Peters BA, Dominianni C, Shapiro JA, Church TR, Wu J, Miller G, Yuen E, Freiman H, Lustbader I, Salik J, Friedlander C, Hayes RB, Ahn J (2016) The gut microbiota in conventional and serrated precursors of colorectal cancer. Microbiome 4(1):69. https://doi.org/10.1186/s40168-016-0218-6
Peterson DA, Frank DN, Pace NR, Gordon JI (2008) Metagenomic approaches for defining the pathogenesis of inflammatory bowel diseases. Cell Host Microbe 3(6):417–427. https://doi.org/10.1016/j.chom.2008.05.001
Planer JD, Peng Y, Kau AL, Blanton LV, Ndao IM, Tarr PI, Warner BB, Gordon JI (2016) Development of the gut microbiota and mucosal IgA responses in twins and gnotobiotic mice. Nature 534(7606):263–266. https://doi.org/10.1038/nature17940
Ramette A (2007) Multivariate analyses in microbial ecology. FEMS Microbiol Ecol 62(2):142–160. https://doi.org/10.1111/j.1574-6941.2007.00375.x
Reeves AE, Koenigsknecht MJ, Bergin IL, Young VB (2012) Suppression of Clostridium difficile in the gastrointestinal tracts of germfree mice inoculated with a murine isolate from the family Lachnospiraceae. Infect Immun 80(11):3786–3794. https://doi.org/10.1128/IAI.00647-12
Rognes T, Flouri T, Nichols B, Quince C, Mahe F (2016) VSEARCH: a versatile open source tool for metagenomics. PeerJ 4:e2584. https://doi.org/10.7717/peerj.2584
Schwabe RF, Jobin C (2013) The microbiome and cancer. Nat Rev Cancer 13(11):800–812. https://doi.org/10.1038/nrc3610
Shah MS, DeSantis TZ, Weinmaier T, McMurdie PJ, Cope JL, Altrichter A, Yamal JM, Hollister EB (2018) Leveraging sequence-based faecal microbial community survey data to identify a composite biomarker for colorectal cancer. Gut 67(5):882–891. https://doi.org/10.1136/gutjnl-2016-313189
Shen H, Ye XY, Li XF, Pan WS, Yuan Y (2015) Severe esophageal bleeding in colorectal cancer due to antitumor therapy: a case report. Oncol Lett 10(6):3660–3662. https://doi.org/10.3892/ol.2015.3742
Shi X, Wong YD, Li MZ, Palanisamy C, Chai C (2019) A feature learning approach based on XGBoost for driving assessment and risk prediction. Accid Anal Prev 129:170–179. https://doi.org/10.1016/j.aap.2019.05.005
Siegel RL, Miller KD, Fuchs HE (2021) Jemal A (2021) Cancer statistics. CA Cancer J Clin 71(1):7–33. https://doi.org/10.3322/caac.21654
Siegel RL, Miller KD, Goding Sauer A, Fedewa SA, Butterly LF, Anderson JC, Cercek A, Smith RA (2020) Jemal A (2020) Colorectal cancer statistics. CA Cancer J Clin 70(3):145–164. https://doi.org/10.3322/caac.21601
Tahara T, Yamamoto E, Suzuki H, Maruyama R, Chung W, Garriga J, Jelinek J, Yamano HO, Sugai T, An B, Shureiqi I, Toyota M, Kondo Y, Estecio MR, Issa JP (2014) Fusobacterium in colonic flora and molecular features of colorectal carcinoma. Cancer Res 74(5):1311–1318. https://doi.org/10.1158/0008-5472.CAN-13-1865
Triantafillidis JK, Vagianos C, Malgarinos G (2015) Colonoscopy in colorectal cancer screening: current aspects. Indian J Surg Oncol 6(3):237–250. https://doi.org/10.1007/s13193-015-0410-3
Varghese C, Shin HR (2014) Strengthening cancer control in China. Lancet Oncol 15(5):484–485. https://doi.org/10.1016/S1470-2045(14)70056-7
Vogtmann E, Hua X, Zeller G, Sunagawa S, Voigt AY, Hercog R, Goedert JJ, Shi J, Bork P, Sinha R (2016) Colorectal cancer and the human gut microbiome: reproducibility with whole-genome shotgun sequencing. PLoS ONE 11(5):e0155362. https://doi.org/10.1371/journal.pone.0155362
Voigt AY, Costea PI, Kultima JR, Li SS, Zeller G, Sunagawa S, Bork P (2015) Temporal and technical variability of human gut metagenomes. Genome Biol 16:73. https://doi.org/10.1186/s13059-015-0639-8
Xia Z, Han Y, Wang K, Guo S, Wu D, Huang X, Li Z, Zhu L (2017) Oral administration of propionic acid during lactation enhances the colonic barrier function. Lipids Health Dis 16(1):62. https://doi.org/10.1186/s12944-017-0452-3
Yokono M, Satoh S, Tanaka A (2018) Comparative analyses of whole-genome protein sequences from multiple organisms. Sci Rep 8(1):6800. https://doi.org/10.1038/s41598-018-25090-8
Yu T, Guo F, Yu Y, Sun T, Ma D, Han J, Qian Y, Kryczek I, Sun D, Nagarsheth N, Chen Y, Chen H, Hong J, Zou W, Fang JY (2017) Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell 170(3):548-563 e516. https://doi.org/10.1016/j.cell.2017.07.008
Zackular JP, Rogers MA, Ruffin MTT, Schloss PD (2014) The human gut microbiome as a screening tool for colorectal cancer. Cancer Prev Res (phila) 7(11):1112–1121. https://doi.org/10.1158/1940-6207.CAPR-14-0129
Zeller G, Tap J, Voigt AY, Sunagawa S, Kultima JR, Costea PI, Amiot A, Bohm J, Brunetti F, Habermann N, Hercog R, Koch M, Luciani A, Mende DR, Schneider MA, Schrotz-King P, Tournigand C, Tran Van Nhieu J, Yamada T, Zimmermann J, Benes V, Kloor M, Ulrich CM, von Knebel DM, Sobhani I, Bork P (2014) Potential of fecal microbiota for early-stage detection of colorectal cancer. Mol Syst Biol 10:766. https://doi.org/10.15252/msb.20145645
Zhang Y, Shen J, Shi X, Du Y, Niu Y, Jin G, Wang Z, Lyu J (2021) Gut microbiome analysis as a predictive marker for the gastric cancer patients. Appl Microbiol Biotechnol 105(2):803–814. https://doi.org/10.1007/s00253-020-11043-7
Zhou Y, Li K, Du Y, Wu Z, Wang H, Zhang X, Yang Y, Chen L, Hao K, Wang Z, Lyu J (2021) Protein interacting with C-kinase 1 is involved in epithelial-mesenchymal transformation and suppresses progress of gastric cancer. Med Oncol 38(4):34. https://doi.org/10.1007/s12032-021-01483-0
Funding
This research was supported by Zhejiang Provincial Natural Science Foundation of China (grant no. LQ21H200007) and Zhejiang Provincial Medical and Health Science and Technology Project of China (grant nos. 2017KY216 and 2017KY486).
Author information
Authors and Affiliations
Contributions
YQD, YFN, and XWS conceived and designed this study. JS, GLJ, and ZLZ collected and arranged the clinical samples. JS, XWS, and JZ performed the analysis of the clinical data. JS, YS, and YZZ prepared 16S rRNA gene amplicons for sequencing. Bioinformatics analyses and statistical analyses were done by GLJ, YQD, XXX, and TTM. All authors contributed to the manuscript preparation and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval
Ethical approval and experimental protocols were supported by Zhejiang Provincial People’s Hospital in Hangzhou, Zhejiang, China (No. 2021QT026). Informed consent had been obtained from all of the participants in this study.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Shen, J., Jin, G., Zhang, Z. et al. A multiple-dimension model for microbiota of patients with colorectal cancer from normal participants and other intestinal disorders. Appl Microbiol Biotechnol 106, 2161–2173 (2022). https://doi.org/10.1007/s00253-022-11846-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-022-11846-w