Abstract
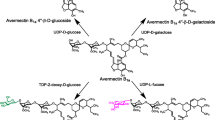
Avermectin, produced by Streptomyces avermitilis, is an active compound protective against nematodes, insects, and mites. However, its potential usage is limited by its low aqueous solubility. The uridine diphosphate (UDP)-glycosyltransferase (BLC) from Bacillus licheniformis synthesizes avermectin glycosides with improved water solubility and in vitro antinematodal activity. However, enzymatic glycosylation of avermectin by BLC is limited due to the low conversion rate of this reaction. Thus, improving BLC enzyme activity is necessary for mass production of avermectin glycosides for field application. In this study, the catalytic activity of BLC toward avermectin was enhanced via directed evolution. Three mutants from the BLC mutant library (R57H, V227A, and D252V) had specific glucosylation activity for avermectin 2.0-, 1.8-, and 1.5-fold higher, respectively, than wild-type BLC. Generation of combined mutations via site-directed mutagenesis led to even further enhancement of activity. The triple mutant, R57H/V227A/D252V, had the highest activity, 2.8-fold higher than that of wild-type BLC. The catalytic efficiencies (Kcat/Km) of the best mutant (R57H/V227A/D252V) toward the substrates avermectin and UDP-glucose were improved by 2.71- and 2.29-fold, respectively, compared to those of wild-type BLC. Structural modeling analysis revealed that the free energy of the mutants was − 1.1 to − 7.1 kcal/mol lower than that of wild-type BLC, which was correlated with their improved activity.
Key points
• Directed evolution improved the glucosylation activity of BLC toward avermectin.
• Combinatorial site-directed mutagenesis led to further enhanced activity.
• The mutants exhibited lower free energy values than wild-type BLC.


Similar content being viewed by others
References
Ahn BC, Kim BG, Jeon YM, Lee EJ, Lim Y, Ahn JH (2009) Formation of flavone di-O-glucosides using a glycosyltransferase from Bacillus cereus. J Microbiol Biotechnol 19(4):387–390
Bolam DN, Roberts S, Proctor MR, Turkenburg JP, Dodson EJ, Martinez-Fleites C, Yang M, Davis BG, Davies GJ, Gilbert HJ (2007) The crystal structure of two macrolide glycosyltransferases provides a blueprint for host cell antibiotic immunity. Proc Natl Acad Sci U S A 104(13):5336–5341. https://doi.org/10.1073/pnas.0607897104
Bowles D, Lim EK, Poppenberger B, Vaistij FE (2006) Glycosyltransferases of lipophilic small molecules. Annu Rev Plant Biol 57:567–597. https://doi.org/10.1146/annurev.arplant.57.032905.105429
Burg RW, Miller BM, Baker EE, Birnbaum J, Currie SA, Hartman R, Kong Y-L, Monaghan RL, Olson G, Putter I, Tunac JB, Wallick H, Stapley EO, Oiwa R, Ōmura S (1979) Avermectins, new family of potent anthelmintic agents: producing organism and fermentation. Antimicrob Agents Chemother 15(3):361–367
Buss O, Rudat J, Ochsenreither K (2018) FoldX as protein engineering tool: better than random based approaches? Comput Struct Biotechnol J 16:25–33. https://doi.org/10.1016/j.csbj.2018.01.002
Chandler PG, Broendum SS, Riley BT, Spence MA, Jackson CJ, McGowan S, Buckle AM (2020) Strategies for increasing protein stability. Methods Mol Biol 2073:163–181. https://doi.org/10.1007/978-1-4939-9869-2_10
Choi HY, Kim BM, Morgan AMA, Kim JS, Kim WG (2017) Improvement of the pharmacological activity of menthol via enzymatic beta-anomer-selective glycosylation. AMB Express 7(1):167. https://doi.org/10.1186/s13568-017-0468-0
Choi H-Y, Van Minh N, Choi JM, Hwang JY, Seo S-T, Lee S-K, Kim W-G (2018) Enzymatic synthesis of avermectin B 1a glycosides for the effective prevention of the pine wood nematode Bursaphelenchus xylophilus. Appl Microbiol Biotechnol 102(5):2155–2165
Conway P, Tyka MD, DiMaio F, Konerding DE, Baker D (2014) Relaxation of backbone bond geometry improves protein energy landscape modeling. Protein Sci 23(1):47–55. https://doi.org/10.1002/pro.2389
Costanzo JA, O’Brien CJ, Tiller K, Tamargo E, Robinson AS, Roberts CJ, Fernandez EJ (2014) Conformational stability as a design target to control protein aggregation. Protein Eng Des Sel 27(5):157–167. https://doi.org/10.1093/protein/gzu008
Cui B, Wang C, Zhao X, Yao J, Zeng Z, Wang Y, Sun C, Liu G, Cui H (2018) Characterization and evaluation of avermectin solid nanodispersion prepared by microprecipitation and lyophilisation techniques. PLoS One 13(1): e0191742. https://doi.org/10.1371/journal.pone.0191742
Denard CA, Ren H, Zhao H (2015) Improving and repurposing biocatalysts via directed evolution. Curr Opin Chem Biol 25:55–64. https://doi.org/10.1016/j.cbpa.2014.12.036
Egerton JR, Ostlind DA, Blair LS, Eary CH, Suhayda D, Cifelli S, Riek RF, Campbell WC (1979) Avermectins, new family of potent anthelmintic agents: efficacy of the B1a component. Antimicrob Agents Chemother 15(3):372–378. https://doi.org/10.1128/aac.15.3.372
Gianese G, Bossa F, Pascarella S (2003) Improvement in prediction of solvent accessibility by probability profiles. Protein Eng 16(12):987–992. https://doi.org/10.1093/protein/gzg139
Guo J, Ma R, Su B, Li Y, Zhang J, Fang J (2016) Raising the avermectins production in Streptomyces avermitilis by utilizing nanosecond pulsed electric fields (nsPEFs). Sci Rep 6:25949
Hu S, Huang J, Mei L, Yu Q, Yao S, Jin Z (2010) Altering the regioselectivity of cytochrome P450 BM-3 by saturation mutagenesis for the biosynthesis of indirubin. J Mol Catal B Enzym 67(1-2):29–35
Huang B, Zhao Q, Zhou JH, Xu G (2019) Enhanced activity and substrate tolerance of 7alpha-hydroxysteroid dehydrogenase by directed evolution for 7-ketolithocholic acid production. Appl Microbiol Biotechnol 103(6):2665–2674. https://doi.org/10.1007/s00253-019-09668-4
Ikeda H, Ōmura S (1997) Avermectin biosynthesis. Chem Rev 97(7):2591–2610
Jones HBL, Wells SA, Prentice EJ, Kwok A, Liang LL, Arcus VL, Pudney CR (2017) A complete thermodynamic analysis of enzyme turnover links the free energy landscape to enzyme catalysis. FEBS J 284(17):2829–2842. https://doi.org/10.1111/febs.14152
Kather I, Jakob RP, Dobbek H, Schmid FX (2008) Increased folding stability of TEM-1 beta-lactamase by in vitro selection. J Mol Biol 383(1):238–251. https://doi.org/10.1016/j.jmb.2008.07.082
Kellogg EH, Leaver-Fay A, Baker D (2011) Role of conformational sampling in computing mutation-induced changes in protein structure and stability. Proteins 79(3):830–838. https://doi.org/10.1002/prot.22921
Khersonsky O, Rosenblat M, Toker L, Yacobson S, Hugenmatter A, Silman I, Sussman JL, Aviram M, Tawfik DS (2009) Directed evolution of serum paraoxonase PON3 by family shuffling and ancestor/consensus mutagenesis, and its biochemical characterization. Biochem 48(28):6644–6654. https://doi.org/10.1021/bi900583y
Liu Z, Lemmonds S, Huang J, Tyagi M, Hong L, Jain N (2018) Entropic contribution to enhanced thermal stability in the thermostable P450 CYP119. Proc Nat Acad Sci U S A 115(43):E10049–E10058. https://doi.org/10.1073/pnas.1807473115
Marabotti A, Scafuri B, Facchiano A (2020) Predicting the stability of mutant proteins by computational approaches: an overview. Brief Bioinform. https://doi.org/10.1093/bib/bbaa074
Navratilova V, Paloncyova M, Berka K, Mise S, Haga Y, Matsumura C, Sakaki T, Inui H, Otyepka M (2017) Molecular insights into the role of a distal F240A mutation that alters CYP1A1 activity towards persistent organic pollutants. Biochim Biophys Acta Gen Subj 1861(11 Pt A):2852–2860. https://doi.org/10.1016/j.bbagen.2017.08.002
Ostlind D, Cifelli S, Lang R (1979) Insecticidal activity of the anti-parasitic avermectins. Vet Rec 105(8):168–168. https://doi.org/10.1136/vr.105.8.168-a
Pandey RP, Gurung RB, Parajuli P, Koirala N, Tuoi le T, Sohng JK (2014) Assessing acceptor substrate promiscuity of YjiC-mediated glycosylation toward flavonoids. Carbohydr Res 393:26–31. https://doi.org/10.1016/j.carres.2014.03.011
Putter I, Connell JGM, Preiser FA, Haidri AA, Ristich SS, Dybas RA (1981) Avermectins: novel insecticides, acaricides and nematicides from a soil microorganism. Experientia 37(9):963–964. https://doi.org/10.1007/bf01971780
Rose GD, Geselowitz AR, Lesser GJ, Lee RH, Zehfus MH (1985) Hydrophobicity of amino acid residues in globular proteins. Sci 229(4716):834–838. https://doi.org/10.1126/science.4023714
Shao H, He X, Achnine L, Blount JW, Dixon RA, Wang X (2005) Crystal structures of a multifunctional triterpene/flavonoid glycosyltransferase from Medicago truncatula. Plant Cell 17(11):3141–3154. https://doi.org/10.1105/tpc.105.035055
Socha RD, Tokuriki N (2013) Modulating protein stability-directed evolution strategies for improved protein function. FEBS J 280(22):5582–5595. https://doi.org/10.1111/febs.12354
Strub C, Alies C, Lougarre A, Ladurantie C, Czaplicki J, Fournier D (2004) Mutation of exposed hydrophobic amino acids to arginine to increase protein stability. BMC Biochem 5:9. https://doi.org/10.1186/1471-2091-5-9
Taylor JL, Price JE, Toney MD (2015) Directed evolution of the substrate specificity of dialkylglycine decarboxylase. Biochim Biophys Acta 1854(2):146–155. https://doi.org/10.1016/j.bbapap.2014.12.003
Tian K, Tai K, Chua BJW, Li Z (2017) Directed evolution of Thermomyces lanuginosus lipase to enhance methanol tolerance for efficient production of biodiesel from waste grease. Bioresour Technol 245(Pt B):1491–1497. https://doi.org/10.1016/j.biortech.2017.05.108
Turner NJ (2009) Directed evolution drives the next generation of biocatalysts. Nat Chem Biol 5(8):567–573. https://doi.org/10.1038/nchembio.203
Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, de Beer TAP, Rempfer C, Bordoli L, Lepore R, Schwede T (2018) SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res 46(W1):W296–W303. https://doi.org/10.1093/nar/gky427
Weymouth-Wilson AC (1997) The role of carbohydrates in biologically active natural products. Nat Prod Rep 14(2):99–110
Wu CZ, Jang JH, Woo M, Ahn JS, Kim JS, Hong YS (2012) Enzymatic glycosylation of nonbenzoquinone geldanamycin analogs via Bacillus UDP-glycosyltransferase. Appl Environ Microbiol 78(21):7680–7686. https://doi.org/10.1128/AEM.02004-12
Yasutake Y, Kameda T, Tamura T (2017) Structural insights into the mechanism of the drastic changes in enzymatic activity of the cytochrome P450 vitamin D3 hydroxylase (CYP107BR1) caused by a mutation distant from the active site. Acta Crystallogr F Struct Biol Commun 73(Pt 5):266–275. https://doi.org/10.1107/S2053230X17004782
Availability of data and materials
The datasets supporting the conclusions of this article are included within the article and its supplementary information file.
Funding
This study was supported by grants from the National Institute of Forest Science (Project No. FE0702-2016-11-2020) and the KRIBB Research Initiative Program (KGM2112133), Republic of Korea.
Author information
Authors and Affiliations
Contributions
WGK conceived and designed research. HYC and HSL performed experiments and analyzed data. KHP conducted structural modeling. JHK contributed reagents, review, and editing. HYC, KHP, and WGK wrote the paper. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
ESM 1
(PDF 506 kb)
Rights and permissions
About this article
Cite this article
Choi, HY., Lim, H.S., Park, KH. et al. Directed evolution of glycosyltransferase for enhanced efficiency of avermectin glucosylation. Appl Microbiol Biotechnol 105, 4599–4607 (2021). https://doi.org/10.1007/s00253-021-11279-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-021-11279-x