Abstract
Shigellosis is a significant type of diarrhea that causes 160,000 deaths annually in a global scale. The mortality occurs mainly in children less than 5 years of age. No licensed vaccine is available, and conventional efforts for developing an effective and safe vaccine against shigellosis have not been succeeded yet. The reverse vaccinology is a novel promising method that screens genome or proteome of an organism for finding new vaccine candidates. In this study, through reverse vaccinology approach, new vaccine candidates against Shigella flexneri were identified and experimentally evaluated. Proteomes of S. flexneri were obtained from UniProt, and then outer membrane and extracellular proteins were predicted and selected for the evaluation of transmembrane domains, protein conservation, host homology, antigenicity, and solubility. From 103 proteins, 7 high-scored proteins were introduced as novel vaccine candidates, and after B- and T-cell epitope prediction, the best protein was selected for experimental studies. Recombinant protein was expressed, purified, and injected to BALB/c mice. The adhesion inhibitory effect of sera was also studied. The immunized mice demonstrated full protection against the lethal dose challenge. The sera remarkably inhibited S. flexneri adhesion to Caco-2 epithelial cells. The results indicate that identified antigen can serve for vaccine development against shigellosis and support reverse vaccinology for discovering novel effective antigens.
Key points
• Seven Shigella new antigens were identified by reverse vaccinology (RV) approach.
• The best antigen experimented demonstrated full protection against lethal dose.
• In vivo results verified RV analyses and suggest FimG as a new potent vaccine candidate.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Diarrhea is a global health-threatening disease that causes approximately 1.3 million deaths annually and has the second mortality rate in children under 5 years old, mostly in developing countries. Terribly, each year 500,000 young children are deceased by diarrhea, and Shigella is one of the major component to this burden (Chen and Kotloff 2016; Bakera and Thea 2018). The Global Enteric Multicenter Study (GEMS), from 22 diarrheal pathogens, considered rotavirus, Shigella, enterotoxigenic Escherichia coli (ETEC), and Cryptosporidium responsible for 70% of moderate-to-severe (MSD) diarrhea cases in 0–4-year-olds (Anderson et al. 2019a). World Health Organization (WHO) on African and the Eastern Mediterranean regions reported ETEC and Shigella as substantial global health problems (Anderson et al. 2019b).
Shigellosis or bacillary dysentery is a type of bacterial diarrhea caused by gram-negative genus Shigella, especially in developing countries. Shigellosis causes acute inflammation of the distal sections of the colon and the rectum (Okafor 2018), leading to 160,000 deaths annually, of which one-third are young children (Bakera and Thea 2018). Shigella is transmitted by fecal-oral route and has a low infectious dose of 10–100 bacteria to initiate disease (Chitradevi et al. 2016). Most of the shigellosis cases occur in low hygienic regions through infected water and food or direct personal contacts. The infants and aged ones are the most vulnerable groups to shigellosis (Okafor 2018).
Shigella is a pathovar of Escherichia coli including four serogroups with more than 50 serotypes (Anderson et al. 2016; Chitradevi et al. 2016). Based on the diversity of O-antigen structures of outer membrane lipopolysaccharide (LPS), Shigella is categorized to serogroup A: Shigella dysenteriae (12 serotypes), Serogroup B: Shigella flexneri (6 serotypes), Serogroup C: Shigella boydii (23 serotypes), and Serogroup D: Shigella sonnei (1 serotype) (Okafor 2018). S. flexneri is the widespread species around the world and in charge of most cases in the developing nations, but S. sonnei is more prevalent in industrialized states and developed countries (Kotloff et al. 2013).
The infection cycle of Shigella begins with bacterial invasion to the colonic epithelium. Following the entrance of bacteria to the small intestine, the early symptoms of diarrhea come up (Ashida et al. 2013), but Shigella mainly affects the colonic epithelium and leads to main symptoms like fever, abdominal cramps, and bloody or mucoid diarrhea (Chitradevi et al. 2016). To overcome host microbiota, Shigella secretes colicin, a small cytotoxic protein. In addition, mucus combination altered by enzymes such as mucinase and neuraminidase is produced by all 4 Shigella species (Anderson et al. 2016). The three main steps of Shigella pathogenesis include adherence to the host cell, invasion and entry to epithelial cells, and spread within the colonic epithelium (Mattock and Blocker 2017). A 220-kb virulence plasmid has an important role in Shigella pathogenesis (Morris et al. 2013). Most of the Shigella virulence factors are located in 20–30 kb part of virulence plasmid, termed the “entry region.” The type III secretion system (T3SS) is a macromolecular needle-shaped complex that is necessary for invasion and escape of autophagy and is encoded by entry region. In addition, T3SS delivers effectors to host cell cytoplasm to manipulate cellular activities and induce apoptosis (Mattock and Blocker 2017).
For treatment of shigellosis, the antibiotic therapy can be effective. Ciprofloxacin is the first-line prescribed antibiotic, and the second preferences are pivmecillinam and ceftriaxone. Most of these antibiotics are expensive and with no pediatric formulation (Williams and Berkley 2018). Also, due to the emergence of globally antibiotic resistance against Shigella, preventive measures are needed. Currently, there is no licensed vaccine against shigellosis, while numerous vaccine candidates are in different trial steps (Frenck Jr et al. 2018; Gilavand et al. 2020; Medeiros et al. 2020).
In recent years, improvement of numerous databases, and development of a plethora of in silico algorithms and statistical methods, expanded the sequence-based approaches in vaccine research and development (Schussek et al. 2014). Reverse vaccinology (RV) is a systematic detection of entire potential antigens for developing a theoretically effective and safe vaccine against pathogens (Mora et al. 2003). Reverse vaccinology, unlike traditional vaccinology, virtually identifies all putative antigens and also uncultivable pathogens. In addition in RV, among the novel identified antigens, the most conserved and immunogenic vaccine candidates can be selected (Sette and Rappuoli 2010).
The first successful experience of reverse vaccinology approach was used for the detection of vaccine candidates against Neisseria meningitidis serogroup B (MenB) and led into a universal MenB vaccine (Bambini and Rappuoli 2009). This achievement encouraged the researchers to use the approach for a variety of other pathogens such as Staphylococcus aureus (Oprea and Antohe 2013), Pseudomonas aeruginosa (Rashid et al. 2017), Streptococcus pneumoniae (Talukdar et al. 2014), Acinetobacter baumannii (Chiang et al. 2015), Helicobacter pylori (Naz et al. 2015), Brucella melitensis (Vishnu et al. 2017), Rickettsia prowazekii (Caro-Gomeza et al. 2014), and Histophilus somni (Madampage et al. 2015).
An efficient Shigella vaccine must be prepared for multiple serotypes to induce broad-spectrum immunity. The identified vaccine candidate should also provoke both systemic and mucosal immune systems (Okafor 2018). In this study, in order to identify novel conserved antigens for vaccine development, high-throughput screening of S. flexneri surface proteins was performed. The immunogenicity and protectivity of the new vaccine candidate were verified experimentally.
Material and methods
Subcellular localization and topology evaluation
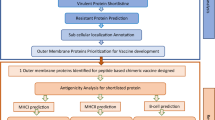
Proteomes of S. flexneri 2a were retrieved from UniProtKB in FASTA format and then imported to CELLO v.2.5 and PSORTb v.3.0.2 online servers for subcellular localization. Surface-exposed proteins, due to their close contacts with host cells, and hence immune system stimulations are appropriate subunit vaccine targets. Thus, in this study and as a starting step, the outer membrane and extracellular proteins that were predicted in both CELLO and PSORTb programs were selected. PSORTb v.3.0 is a precise and wide-used online program that utilizes multiple analytical modules for the identification of known biological features related to proteins’ subcellular locations (Yu et al. 2010). Query proteins, which were predicted as outer membrane or extracellular proteins with probability value more than 0.9 (cut-off 9), were selected. CELLO is based on multi-class support vector machines (SVMs) and has a prediction accuracy of 89% localized bacterial proteins (Yu et al. 2006). In CELLO, predicted outer membrane or extracellular proteins with the highest reliability score were selected for subsequent analyses.
Due to difficulty in cloning and expression of proteins with transmembrane domains > 1, these proteins were identified and ruled out. The TMHMM v.2.0 by searching hydrophobic amino acids and HMMTOP v.2.0 with a maximum divergence of amino acid composition, using default values of their parameters, were used to determine the number of transmembrane domains in outer membrane and extracellular proteins (Krogh et al. 2001; Tusnády and Simon 2001).
Homology analyses
To prevent auto-immune response, the homology of selected proteins with human (taxid: 9606) and mouse (taxid: 10088) proteins was evaluated by the online BLAST server, provided by NCBI database (Talukdar et al. 2014). BLASTp was performed against non-redundant protein sequences, and proteins with identity and query cover > 30% were defined as homologous proteins and were eliminated (blast was performed with default values of their parameters) (Altschul et al. 1990; Rashid et al. 2017). An applicable vaccine candidate should be protective against different species and strains, to be used as a broad-spectrum vaccine (Kim et al. 2015), and therefore, the determination of protein conservation is necessary. To determine orthologous proteins, FASTA sequences aligned against non-redundant proteins of genus Shigella (taxid:620), and proteins with identity and query cover more than 80% and with E-value threshold of 10−6 in all 4 species of Shigella were selected.
Antigenicity and solubility prediction
FASTA sequences of preselected proteins were submitted to VaxiJen server for the prediction of protein antigenicity. VaxiJen, with an alignment-free method using only the physiochemical properties of amino acid sequences, is the first software that predicts antigenicity via machine learning method (Flower et al. 2017). To increase the reliability of antigenicity prediction, the ANTIGENpro online tool, which uses a large non-redundant microarray dataset, was employed to predict antigenicity (Magnan et al. 2010). Proteins with prediction scores more than 0.7 in both VaxiJen and ANTIGENpro were selected as probable antigens, and then, the solubility of these proteins was predicted. Efficient expression of heterologous proteins in E. coli leads to the successful production of protein in active form. PROSO II is an online software that employs two-layered classification method and Parzen window model for the prediction of protein solubility (Smialowski et al. 2012). Protein-Sol is also a web tool that by sequence-based properties like amino acid compositions, isoelectric pH, fold propensity, and sequence entropy predicts protein solubility (Hebditch et al. 2017). Proteins with calculated scores more than 0.6 and 0.45 in PROSO II and Protein-Sol, respectively, were considered as soluble proteins and listed for next analyses.
Epitope mapping
The shortlisted proteins from the above analyses, after literature screening, were introduced as potential novel antigens, and for the selection of the best protein for experimental studies, B- and T-cell epitopes were predicted. To increase prediction reliability, different programs with different methods were used. Immune Epitope Database (IEDB) based on experimental data (Vita et al. 2019), BCPred by support vector machine (SVM) method (El-Manzalawy et al. 2008), BcePred using different physicochemical properties (Saha and Raghava 2004), and ABCpred based on the recurrent neural network method (Saha and Raghava 2006) with default values of their parameters were used for the calculation of new antigens’ epitope density (the ratio between the summation of B-epitope scores and the protein length) (Oprea and Antohe 2013; Meunier et al. 2016). T cells are the other part of the adaptive immune system and have a crucial role in human immunogenicity. In IEDB database, high-affinity binding epitopes with IC50 and percentile rank < 5 nm and for full HLA reference set (54 alleles in MHC-I and 27 alleles in MHC-II) were determined. Also, NetMHC 4.0, which utilizes artificial neural network method, was used for the prediction of strong binders (percent rank < 0.5) for all the 81 human MHC alleles with 8–14 amino acid length (Andreatta and Nielsen 2016). MHCPred v.2.0, based on a quantitative structure-activity relationship (QSAR) method, is another T-epitope prediction tool (Hattotuwagama et al. 2004) and was used for the prediction of strong binders’ T-cell epitopes (IC50 < 50 nm) in all 25 alleles, which were provided by the server. Shigella belongs to intracellular bacterial pathogens and provokes cytotoxic T lymphocytes (CTLs) (Cossart and Helenius 2014; Mak and Saunders 2006). Therefore, NetCTL v.1.2 was employed for CTL epitope prediction on 12 MHC super types, with a threshold value of 0.75 (Larsen et al. 2007). Then, T-epitope density was calculated as the number of high-affinity-predicted T-epitopes divided by protein length. The results of epitope prediction for selected proteins, in addition to antigenicity and solubility scores, were listed and ranked, and ultimately, the best protein with the overall highest score (in different methods) was selected and characterized for the experimental phase.
Characterization of newly identified vaccine candidate
For the examination of virulence function of selected protein, VirulentPred online server, by means of cascade support vector machine, was used (Garg and Gupta 2008). The MOTIF is one of the GenomeNet database tools that, by searching sequence similarity against major motif libraries, determines protein motifs from amino acid sequence (Kanehisa et al. 2002). ProtParam is one of the online ExPASy tools and, on the basis of pK and N-end rule, determines the special properties of proteins (Gasteiger et al. 2005). SignalP 5.0 server based on deep neural network method predicts signal peptide (SP) sequences and discriminates between three main types of SPs (Almagro Armenteros et al. 2019). Signal sequences are cleaved during protein translocation through the cell membrane (Coleman et al. 1985). Thus, signal peptide sequence was predicted by SignalP server and removed from the complete protein sequence. The website addresses of the servers used in this article are presented in Table 1.
Bacterial strains and growth conditions
S. flexneri (DDBCC1000, http://www.wfcc.info/ccinfo/collection/by_id/1185), E. coli BL21-DE3, and E. coli DH5α were obtained from the bacterial collection center of research division of Shahed University, Tehran, Iran. The virulence potency of the S. flexneri was previously verified by the Sereny test (Sereny 1955). All bacterial strains were grown in a Luria-Bertani (LB) medium at 37 °C. Bacteria containing recombinant plasmids were cultured by the addition of ampicillin (100 μg/mL) or kanamycin (70 μg/mL) to the growth medium.
Gene cloning and protein expression
After in silico analyses and selection of the best protein as a vaccine candidate, the selected gene was optimized by Gene Designer software (https://www.atum.bio/eCommerce/login) according to codon bias of E. coli and synthesized in pUC57 vector (Biomatic, Canada). The gene was subcloned into pET-28a and transferred into E. coli BL21-DE3.
Protein expression was induced by the auto-induction method (Studier 2005). After 18 h of incubation at 37 °C, the bacteria were harvested by centrifugation at 4000×g for 30 min. The pellet was suspended in a Tris-EDTA (TE) buffer and lysed by an ultrasonic disintegrator at ice-cold temperature and intermittent bursts of 1 min. The cell lysate was centrifuged at 10,000×g for 20 min, and the pellet was washed in a TE buffer and recollected. The pellet was finally solubilized in 8 mM urea and centrifuged at 10,000×g for 20 min and supernatant was collected. As a negative control, all the above steps were carried out for an uninduced BL21 harboring recombinant plasmid. Fractions were analyzed on SDS-PAGE for protein expression.
Protein purification was carried out by Ni-NTA affinity chromatography. The column was eluted with different concentrations of imidazole, and fractions were analyzed on SDS-PAGE and verified with western blotting using HRP-conjugated anti-histidine antibody. The concentration of purified protein was estimated with the Bradford method (Bradford 1976).
Immunization assay
The female BALB/c mice (5–6 weeks old) were procured from Razi Institute, Karaj, Iran, and maintained under standard unstressed and pathogen-free conditions at an animal care center at Shahed University. Test and control groups of mice (n = 5/group) were injected intraperitoneally (IP) on days 0, 14, and 28. Test mice were immunized with 20 μg of pure protein emulsified with complete Freund’s adjuvant (Sigma), and the control group received only PBS and adjuvant. In the subsequent inoculations, incomplete Freund’s adjuvant was administered in the same way. One week after each injection, blood was collected by puncturing of the supraorbital plexus in both test and control groups, and blood sera were stored at − 20 °C for further analyses.
Immunogenicity assay (ELISA)
The antibody response to injected protein was examined by indirect ELISA. Moreover, 100 μL coating buffer containing 2 μg of purified protein was added to 96-well micro-titer plates in duplicate repeats and incubated for 16 h at 4 °C. The plate was incubated in 37 °C. Wells were washed three times with 400 μL of PBST (PBS containing 0.05% Tween 20). The wells were blocked with 150 μL of 5% skim milk in PBS for 1 h. Wells were washed and added with serial dilutions of serum in PBST (1:250 to 1:64000), and after 2 h of incubation at 37 °C, wells were washed, and 100 μL of diluted (1/15000) HRP-conjugated anti-mouse IgG was added to wells and incubated for 2 h. TMB (100 μL) as a substrate was added to wells, and the reaction was terminated with 3N H2SO4 after color development. The optical density was quantified at 450 nm using TECAN ELISA reader. To evaluate the accuracy of the process, two types of negative control were applied (Ag−, without protein and only coating buffer, and Ab−, without serum and only PBST).
The bacteria whole-cell ELISA was also performed with 106 CFU/mL active S. flexneri suspended in 100 μL coating buffer and placed in micro-plate wells. The other steps were performed similarly to that of indirect ELISA aforementioned.
Protectivity challenge
LD50 determination
A total of 100 μL of the overnight cultivated bacteria was added to 100 mL LB broth medium and incubated under steady conditions. The optical density of growing bacteria was calculated at different times at 600 nm and simultaneously diluted and cultured on an LB agar. All the steps were repeated three times under completely sterile conditions. Finally, the standard growth curve was prepared by plotting the optical density against bacteria viable count.
The 50% lethal dose (LD50) was determined by intraperitoneal injection of different doses of bacteria (105–109) to groups of adult female BALB/c mice (n = 7/group).
Active immunity experiments
From an overnight LB culture, 100 μL was added to 100 mL LB broth and incubated to grow to mid-log phase. Bacterial cells were harvested by centrifugation at 8000×g, for 20 min, and the pellets were washed with PBS to remove impurities. The resulted sediment was suspended exactly in PBS to adjust inoculum concentration. Furthermore, 100 μL of three doses corresponding to 2 × LD50, 4 × LD50, and 6 × LD50 was intraperitoneally administered to the test and control groups of vaccinated mice (n = 5/group). Mortality and morbidity were monitored and recorded daily for 1 week after the challenge.
Passive immunity experiments
The sera stored at − 20 °C were thawed in room temperature, and the complement system was inactivated by keeping them under 56 °C for 30 min. Moreover, 100 μL sera from the test and control mice groups were injected intravenously to 5–6-week-old female mice (n = 5/group). After 3 hours, 100 μL inoculum, equivalent to 2 × LD50 bacteria, was injected intraperitoneally and mortality in the mice was recorded for 1 week.
Adhesion assay
The inhibitory effect of sera on bacterial adhesion to epithelial cells was evaluated by Caco-2 human colon epithelial cell lines. Caco-2 cells were cultured in a DMEM medium containing 10% fetal bovine serum (FBS) without any antibiotic. Cells were grown in a fresh medium, in a humidified atmosphere with 5% CO2 at 37 °C. After 7–10 days, the culture medium was discarded and cell layer was washed with PBS and detached with 25% trypsin in PBS. Dispersed cells were then harvested and counted by an inverted microscope, and 105 cells were sub-cultured in 24-well tissue culture plates with DMEM containing 10% FBS and 1% penicillin/streptomycin. The plates were incubated for 48 h at the above-mentioned conditions.
The sera complement system was inactivated and diluted in PBS to a final concentration of 1:250 and then added to 107 CFU/mL S. flexneri for 1 h. The Caco-2 cell layer was washed with PBS and then infected with 500 μL of pretreated bacteria (containing 5 × 106 bacteria and 1:250 serum) for 2 h. The Caco-2 cells were detached with 1% (v/v) trypsin-PBS and cultured in LB agar plates. The number of colonies for S. flexneri pretreated with immunized and control mice sera and also S. flexneri without serum were counted, and then, the percentages of bacteria-binding inhibition to Caco-2 cells subjected to immunized and control mice sera were calculated.
Statistical analyses
The GraphPad Prism 8 software was used for statistical analyses. The statistical differences between antibody responses of immunized and non-immunized mice and the significance of differences in inhibition of adhesion assay test were analyzed by Student’s t-test. Experiments were performed in duplicate, and P-values less than 0.05 were considered significant.
Results
Subcellular localization and topology analyses
In UniProt database, there is only 1 reference proteome of S. flexneri that contains 4103 proteins (serotype 2a, strain 301, UP000001006). Standard and full proteome of S. flexneri 2a strain 2457T that is a highly virulent strain and has been extensively used for clinical researches, including 3786 proteins with 99.3% data completeness, was selected (UP000002673). In S. flexneri 2a strain 301, 108 and 76 outer membrane proteins and 107 and 42 extracellular proteins were predicted in CELLO and PSORTb, respectively. Also, in S. flexneri 2a strain 2457T, 129 and 72 outer membrane and 141 and 37 extracellular proteins were predicted in CELLO and PSORTb, respectively. In S. flexneri 2a strain 301, 54 outer membrane and 26 extracellular proteins were predicted by both CELLO and PSORTb, and in S. flexneri 2a strain 2457T, 57 outer membrane and 29 extracellular proteins were predicted by both CELLO and PSORTb. Finally, by removing repetitive proteins in S. flexneri 301 and 2457T strains, overall 103 proteins (69 outer membrane and 34 extracellular proteins) were selected for next analyses (Table 2). Out of these 103 proteins, 4 and 10 proteins with transmembrane domains > 1 were predicted with TMHMM and HMMTOP, respectively, and then removed from the proteins list (Table 2).
Homology analyses
From 89 proteins obtained, 7 proteins with homology to human and mouse by BLASTp were identified and hence removed. Also, 12 non-conserved proteins among Shigella genus species were identified and eliminated, and 70 proteins were remained to be analyzed in the next step (Table 2).
Antigenicity and solubility prediction
Out of 70 proteins, 23 proteins with antigenicity score > 0.7 were predicted by both VaxiJen and ANTIGENpro programs, and from these, 7 proteins were predicted as soluble proteins in both PROSO II and Protein-Sol programs (Table 2). By screening literatures, none of the selected 7 vaccine candidates were used previously for the development of vaccines against shigellosis, and therefore, these proteins were introduced as novel vaccine candidates. The subtractive pathway for the selection of these proteins is explained in Table 3.
Epitope mapping
The detailed results related to epitope prediction are exhibited in Table 4. Out of 13 different servers, minor fimbrial protein G (FimG) in 6 servers achieved the highest score (ANTIGENpro, PROSO, Protein-Sol, BcePred, MHCPred, and IEDB Class-II T-epitope tool) and also in other servers with a high score was better than other proteins and thus selected as the best vaccine candidate for experimental studies (Table 4).
Characterization of newly identified vaccine candidate
FimG was predicted as a virulence factor in the VirulentPred server (score: 1.0163). Submission of FimG FASTA sequence to MOTIF database leads to the identification of bacterial immunoglobulin-like domain in protein structure (Pfam ID: Big_3_5, family accession: PF16640, i-Evalue: 0.022). Also, signal peptide sequence in FimG was identified by SignalP program (1–23 amino acids). Physicochemical properties of FimG protein were determined in ProtParam server, as demonstrated in Table 5.
Protein expression and purification
The FimG gene was optimized (gene accession number: MW_252166) and synthesized in the pUC57 vector (the optimized gene and other data are available in article Online Resource).Then, the gene was subcloned into pET-28a vector and expressed in E. coli BL21-DE3. The expression results were analyzed by 15% SDS-PAGE, and a 17 kd protein band appeared correctly on the gel (Fig. 1a). The protein was purified under denatured condition by Ni-NTA affinity chromatography and confirmed by western blotting with anti-His-tag and HRP antibody (Fig. 1b).
SDS-PAGE (15%) and western blotting results. Recombinant FimG protein containing His-tagged residues was expressed in E. coli BL21 (DE3), followed by purification byaffinity chromatography using Ni-NTA Agarose matrix. Protein expression was evaluated by SDS-PAGE and Coomassie blue staining (a) and was confirmed by western blotting using conjugated antibodies against His tag (b). (Lane 1: negative control result from non-induced bacteria, Lane 2: recombinant protein expression, Lane 3: purified recombinant protein Lane 4: negative control lysate)
Immunogenicity evaluation
The immunogenicity of immunized mice with FimG was examined by ELISA test. Ten days after each immunization, sera were collected and antibody levels were determined. After second and third administrations, the IgG level significantly increased compared to the control group (Fig. 2a). After third immunization, the murine antibody reaction with S. flexneri was also evaluated in whole-cell ELISA and demonstrated significant sera reaction in comparison with control mice (Fig. 2b). ELISA results from immunized mice after 5-month post-immunization periods showed the high stability of produced antibody.
ELISA analyses of antibody response against recombinant FimG protein. a Indirect ELISA: The sera from the test and control mice groups (n = 5/group) were prepared and tested in duplicate. The results indicated that in the second booster, the immunized mice (test group administered with FimG + adjuvant) in comparison with non-immunized mice (control group administered with PBS + adjuvant) produced significantly higher titer of IgG antibody. The error bars represent standard deviation from two separate experiments. b Antibodies raised against recombinant proteins as determined by whole-cell ELISA
Protectivity challenge
After the injection of different doses of S. flexneri to mice groups, LD50 was determined to be 1.5 × 108 CFU/mL and used for survival challenge. Before the injection, no abnormalities and diseases were observed in immunized mice. Intraperitoneal injection of 2-, 4-, and 6-fold LD50 doses of S. flexneri to control and immunized mice resulted in full protection of immune mice up to 4-fold LD50 and 66% protection against 6LD50 (Fig. 3a). Passive immunity in mice with 2 × LD50 led to 65% survival of challenged mice. The bacteria-infected mice were monitored for 1 week for mortality and morbidity and finally euthanized. The results of protectivity experiments are presented in (Fig. 3b).
a Survival plot of actively immunized mice with intraperitoneal administration of recombinant FimG. After 2 weeks of third administration, the mice (n = 5/group) were challenged with 2 × LD50, 4 × LD50, and 6 × LD50 of S. flexneri through intraperitoneal route and continuously monitored for 168 h (7 days). The immunized mice demonstrated full protection against 4 × LD50 dose of bacteria. b Survival plot of passive immunization. The mice (n = 5/group) received 100 μl of immunized or non-immunized mice sera through intravenous route and, after 3 h, intraperitoneally challenged with 2 × LD50 of S. flexneri. The mice were monitored for 168 h post challenge for mortality and morbidity. The passively immunized mice were significantly protected against 2 × LD50 of bacteria (survival rate 66%). Survival rates were analyzed using the non-parametric log-rank test (P < 0.001)
Adhesion assay
The results of S. flexneri adhesion to Caco-2 cells demonstrated that bacterial adhesion was impaired by produced antibodies against recombinant FimG protein. In comparison to control wells that were treated by control mice sera, immunized mice sera inhibited 29.31% of bacterial adhesion to Caco-2 epithelial cell lines (Fig. 4). Also, the flowchart of reverse vaccinology approach, adopted in this study, is demonstrated in Fig. 5.
Adhesion assay results. Column A and B represents the colony numbers of S. flexneri pretreated with immunized and non-immunized mice sera, respectively, and demonstrate the remarkable sera effect on bacteria binding to Caco-2 cell lines (29.31%) (percentage of serum inhibitory effect on S. flexneri adhesion onto Caco-2 cell = (1 - (176/249)) × 100 = 29.31%)
Discussion
Shigellosis with high mortality and increased multi-drug-resistant strains currently has no reliable vaccine, and studies are ongoing for the identification of new vaccine targets. The first effort to identify Shigella vaccine candidates was performed by Ying et al. in 2005. On the basis of immunoproteomics methods, out of 126 marked spots in 2-dimensional electrophoresis, 87 outer membrane (OM) and extracellular (EC) proteins were identified (Ying et al. 2005). Immunoproteomics-based methods were then used by Jennison et al. (Jennison et al. 2006) in 2006 and Pore and Chakrabarti (2016) in 2016. These methods include protein isolation, 2-D electrophoresis, immunoblotting, peptide mass fingerprinting, and sequence analyzing and therefore are time-consuming methods. Advancement of in silico methods and bioinformatic databases now paved the way for the identification of vaccine candidates, more precise than before. In 2016, Maggiore, in an innovative procedure (GMMA), used genetically modified Shigella extremely producing outer membrane vesicles (OMV) and identified 40 surface protein, common in both S. flexneri and S. sonnei (Maggiore et al. 2016). In a recent study of Leow et al. in 2020, outer membrane proteins of S. flexneri by reverse vaccinology method were evaluated and 5 OM proteins as new vaccine candidates were identified and characterized (Leow et al. 2020). In comparison to our work, they only evaluated OM proteins, and the EC proteins that are as important as OM proteins were ignored and not selected in their studies. In addition, using multiple analytical servers with high selection threshold increased the reliability of our study. Above all, our in silico results were verified experimentally. The 5 identified OM proteins by Leow et al. did not qualify our parameters and were eliminated in our subtractive process.
The first step in routine RV approach is subcellular localization of a set of organism proteins or its proteome. Outer membrane (OM) and extracellular (EC) proteins, due to their direct interaction to host cells and immune system stimulation, are significant immunogenic factors that are employed in many vaccine developments, like Lyme disease and pertussis (Pore and Chakrabarti 2013). These surface-exposed proteins play important roles in bacterial pathogenesis like adhesion, invasion, biofilm formation, effector secretion, and cell-to-cell dissemination (Liao et al. 2009). Shigella outer membrane proteins, such as VirG (IcsA) and OmpA, have been studied as vaccine candidates (Barnoy et al. 2010; Pore and Chakrabarti 2013). Also, Ipa as a Shigella extracellular protein is used for the generation of Invaplex vaccine against Shigella (Riddle et al. 2011).In this study, the OM and EC proteins of S. flexneri were bioinformatically predicted and selected. In silico protein subcellular localization methods are established on three main approaches as sequence homology evaluation, signal peptide recognition, and analyses of total protein properties like amino acid composition. PSORTb program based on all of these three approaches and by different analytical modules is a reliable predictive tool (Yu et al. 2006). PSORTb emphasizes specificity (precision) over sensitivity (recall) and has 98.3% accuracy in gram-negative bacterial subcellular localization, but cannot detect multi-location proteins correctly (Yu et al. 2010). Therefore, CELLO as a complementary program (with 89% accuracy) was used for subcellular localization, and proteins, which were predicted in both CELLO and PSORTb programs, were selected for subsequent analyses.
It has been reported previously that among 600 vaccine candidates of N. meningitides B, 250 proteins, with more than 1 transmembrane domain (TMD), had unsuccessful cloning and expression (Talukdar et al. 2014). Thus, from predicted OM and EC proteins, only proteins with TMD ≤ 1 were selected.
Proteins with homology to human proteins induce auto-immune response and were eliminated from our preselected proteins. In studies carried out by Rashid et al. in 2017 (Rashid et al. 2017) and Vishnu et al. in 2017 (Vishnu et al. 2017), homologous proteins were defined as proteins with more than 40% and 35% identity to human proteins, respectively. We eliminated proteins with more than 30% identity and query cover from shortlisted proteins. Shigella genus has more than 40 serotypes, and an effective vaccine must generate immunity to almost all the serotypes. Thus, to develop a broad-spectrum vaccine against a variety of Shigella strains, the conserved proteins in all the Shigella species must be selected. Caro-Gomeza considered proteins with identity and query cover more than 60% as conserved proteins (Caro-Gomeza et al. 2014). Meunier selects homologous proteins with identity and query cover more than 80 and 50%, respectively (Meunier et al. 2016). Here and among Shigella species, proteins with identity and query cover more than 80%, in all species, were selected.
Expression of recombinant protein in E. coli usually leads to insoluble proteins, and solubilization processes can impair protein activity. Thus, only predicted soluble proteins were selected (Esmailnia et al. 2020; Smialowski et al. 2012).
The host immune responses to S. flexneri include innate, humoral, and cellular immunity (Jennison and Verma 2004).The up-regulation of a series of cytokines is the major innate immune response against Shigella (Ashida et al. 2015; Jennison and Verma 2004). Studies demonstrated that in patients suffering from shigellosis, T cells were proliferated against S. flexneri antigens. Humoral immune response plays a key role in protectivity against shigellosis (Jennison and Verma 2004). Therefore B- and T-cell epitopes were predicted to estimate immunogenic potency of 7 newly identified proteins. To compare these 7 proteins, epitope density regarding protein length and summation of epitope scores was defined and calculated. According to protein scores in different programs, FimG acquired the highest overall score and must be validated in experimental trials.
FimG belongs to type 1 fimbriae family (T1f), which is one of the most frequent bacterial adhesins in the Enterobacteriaceae family (Psonis and Thanassi 2019). Wang et al. in 2019 studied the distribution of virulence factors in 15 clinical strains of S. flexneri and demonstrated that pili proteins are highly conserved virulence factors in these strains (Wang et al. 2019). Also, our blast results showed that FimG is conserved in all Shigella species, E. coli (identity > 97% and query cover 100%), and in other Enterobacteriaceae family members. Fimbrial proteins are present in copious numbers per cell and are involved in bacterial adhesion to host cells, invasion, and colonization on specific tissues (Coleman and Smith 2014). During bacterial infection process, fimbria expression increases and leads to connect bacteria for biofilm formation. They are critical virulence factors and are appropriate vaccine targets (Psonis and Thanassi 2019). For instance, FimA and FimH have been employed for vaccine development in E. coli (Asadi Karam et al. 2013; Ma et al. 2018; Pore and Chakrabarti 2013). FimG in VirulentPred, predicted as a virulence factor and MOTIF server results, indicates that FimG has immunoglobulin-like (Ig-like) domain that takes part in bacterial adhesion to epithelial cells. Similarly, this phenomenon was also found in intimin, a critical virulence factor in EHEC and EPEC that belongs to Ig-like family and plays significant role in bacterial attachment to cell surface (Khare et al. 2010; Son et al. 2002). Sera produced against FimG, in comparison to control mice sera, inhibited bacterial binding to Caco-2 epithelial cells. The adhesion assay results exhibited high inhibitory effect of produced antibody against bacterial adhesion to epithelial cells. These results indicate that FimG is a conserved adhesion protein and therefore validate its virulence function. Moreover, the stability of antibody production, 5 months after the last immunization, was studied and verified by indirect ELISA. This result suggests FimG as a durable and conserved virulence factor.
The humoral immune response against Shigella involves both systemic and mucosal responses. Secretory IgA (sIgA) transcytoses into the intestine, binds to the mucosal layer of the epithelium, and prevents bacterial attachment to mucosal coating (Corthésy 2013; Jennison and Verma 2004). Although IgG and IgM are present in Shigella-infected patients’ serum, the role of IgG, and probably IgM, in the protection against Shigella in mice models is reported. IgA-deficient mice were fully protected against Shigella challenge, indicating that IgG or IgM can provide immunity (Jennison and Verma 2004). In this study, after the third administration, the ELISA test with anti-IgG was carried out. The results showed a high antibody titer of the test mice groups compared to the control groups. Also, whole-cell ELISA results revealed the antibody reaction to native FimG protein on bacterial surface. Therefore, it can be implied that both native and in vitro expressed FimG stimulate the immune system almost equally, and protein immunogenic regions are exposable.
The first bottleneck in vaccinology studies is the determination of an appropriate animal model. Human is the only reservoir of Shigella, and no animal model resembles human immune response perfectly. Primate models like Aotus nancymaae have the most similarity to human immune mechanisms. Due to high-cost effect and handling difficulties, primate models are rare models. Sereny test or keratoconjunctivitis model in guinea pig eyes properly imitates Shigella invasion and intracellular dissemination. This model examines early innate immunity and, due to spontaneous recovery of mice after a few days, cannot validate vaccine efficacy and safety (Gregory et al. 2014; Marteyn 2016). Mice are widespread used models in Shigella vaccine studies. Pulmonary pneumonia model by intranasal bacterial injection is a mostly used model. However, the lung environment is significantly different from the colonic lumen. Adult mice also do not develop shigellosis in oral bacterial administration (Barnoy et al. 2017; Marteyn 2016).
It is demonstrated that intraperitoneal injection of Shigella leads to shigellosis and colonic invasion. The dissemination route is not clearly understood. Administration of different doses of virulent Shigella through intranasal and intraperitoneal route led to successful induction of intraperitoneal injection, whereas, in nasal routes, mild inflammatory symptoms were observed (Sharma et al. 2017). Intraperitoneal injection of Shigella also leads to various pathological symptoms (Yang et al. 2014). In our study, the intraperitoneally immunized mice toward 4 × LD50 dose of bacteria showed full protection, and passive immunization brought about 60% survival rate. The higher protection in active immunization over passive immunization could be due to other immune system pathways and effectors present in active immunity and is in accordance with other reports. Taking into account all these in silico and experimental results, FimG is suggested as a novel and potent vaccine candidate.
In conclusion, experimental results exhibit higher efficiency of FimG as a novel vaccine candidate against shigellosis. These results are in compliance with in silico results and verify the accuracy of our subtractive method and totally the reverse vaccinology method. Therefore, the other identified vaccine candidates introduced in this study can be examined for vaccine development. Also, reverse vaccinology approach as a precise and cutting-edge method can be used for the identification of novel vaccine targets against other pathogens.
Data availability
The supplementary data supporting the conclusions of this article are available in Online Resource (ESM.xlsx), containing results of subtractive process and comparative analyses.
References
Almagro Armenteros JJ, Tsirigos KD, Sønderby CK, Petersen TN, Winther O, Brunak S, von Heijne G, Nielsen H (2019) SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat Biotechnol 37(4):420–423. https://doi.org/10.1038/s41587-019-0036-z
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215(3):403–410. https://doi.org/10.1016/s0022-2836(05)80360-2
Anderson M, Sansonetti PJ, Marteyn BS (2016) Shigella diversity and changing landscape: insights for the twenty-first century. Front Cell Infect Microbiol 6(45). https://doi.org/10.3389/fcimb.2016.00045
Anderson JD, Bagamian KH, Muhib F, Baral R, Laytner LA, Amaya M, Wierzba T, Rheingans R (2019a) Potential impact and cost-effectiveness of future ETEC and Shigella vaccines in 79 low- and lower middle-income countries. Vaccine: X 2:100024. https://doi.org/10.1016/j.jvacx.2019.100024
Anderson JD, Bagamian KH, Muhib F, Amaya MP, Laytner LA, Wierzba T, Rheingans R (2019b) Burden of enterotoxigenic Escherichia coli and shigella non-fatal diarrhoeal infections in 79 low-income and lower middle-income countries: a modelling analysis. Lancet Glob Health 7(3):e321–e330. https://doi.org/10.1016/s2214-109x(18)30483-2
Andreatta M, Nielsen M (2016) Gapped sequence alignment using artificial neural networks: application to the MHC class I system. Bioinformatics 32(4):511–517. https://doi.org/10.1093/bioinformatics/btv639
Asadi Karam MR, Oloomi M, Mahdavi M, Habibi M, Bouzari S (2013) Vaccination with recombinant FimH fused with flagellin enhances cellular and humoral immunity against urinary tract infection in mice. Vaccine 31(8):1210–1216. https://doi.org/10.1016/j.vaccine.2012.12.059
Ashida H, Nakano H, Sasakawa C (2013) Shigella IpaH0722 E3 ubiquitin ligase effector targets TRAF2 to inhibit PKC-NF-κB activity in invaded epithelial cells. PLoS Pathog 9(6):e1003409. https://doi.org/10.1371/journal.ppat.1003409
Ashida H, Mimuro H, Sasakawa C (2015) Shigella manipulates host immune responses by delivering effector proteins with specific roles. Front Immunol 6(219). https://doi.org/10.3389/fimmu.2015.00219
Bakera S, Thea HC (2018) Recent insights into Shigella: a major contributor to the global diarrhoeal disease burden. Curr Opin Infect Dis. https://doi.org/10.1097/QCO.0000000000000475
Bambini S, Rappuoli R (2009) The use of genomics in microbial vaccine development. Drug Discov Today 14(5–6):252–260. https://doi.org/10.1016/j.drudis.2008.12.007
Barnoy S, Jeong KI, Helm RF, Suvarnapunya AE, Ranallo RT, Tzipori S, Venkatesan MM (2010) Characterization of WRSs2 and WRSs3, new second-generation virG (icsA)-based Shigella sonnei vaccine candidates with the potential for reduced reactogenicity. Vaccine 28(6):1642–1654. https://doi.org/10.1016/j.vaccine.2009.11.001
Barnoy S, Gancz H, Zhu Y, Honnold CL, Zurawski DV, Venkatesan MM (2017) The Galleria mellonella larvae as an in vivo model for evaluation of Shigella virulence. Gut Microbes 8(4):335–350. https://doi.org/10.1080/19490976.2017.1293225
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. https://doi.org/10.1006/abio.1976.9999
Caro-Gomeza E, Gazia M, Goeza Y, Valbuena G (2014) Discovery of novel cross-protective Rickettsia prowazekii T-cell antigens using a combined reverse vaccinology and in vivo screening approach. Vaccine 32:4968–4976
Chen WH, Kotloff KL (2016) Shigella vaccine development: finding the path of least resistance. Clin Vaccine Immunol 23(12):904–907. https://doi.org/10.1128/cvi.00444-16
Chiang MH, Sung WC, Lien SP, Chen YZ, Lo AF, Huang JH, Kuo SC, Chong P (2015) Identification of novel vaccine candidates against Acinetobacter baumannii using reverse vaccinology. Hum Vaccin Immunother 11(4):1065–1073. https://doi.org/10.1080/21645515.2015.1010910
Chitradevi STS, Kaur G, Sivaramakrishna U, Singh D, Bansal A (2016) Development of recombinant vaccine candidate molecule against Shigella infection. Vaccine 34(44):5376–5383. https://doi.org/10.1016/j.vaccine.2016.08.034
Coleman JP, Smith CJ (2014) Structure and composition of microbes. reference module in biomedical sciences. Elsevier. https://doi.org/10.1016/B978-0-12-801238-3.05318-6
Coleman J, Inukai M, Inouye M (1985) Dual functions of the signal peptide in protein transfer across the membrane. Cell 43(1):351–360. https://doi.org/10.1016/0092-8674(85)90040-6
Corthésy B (2013) Multi-faceted functions of secretory IgA at mucosal surfaces. Front Immunol 4:185–185. https://doi.org/10.3389/fimmu.2013.00185
Cossart P, Helenius A (2014) Endocytosis of viruses and bacteria. Cold Spring Harb Perspect Biol 6(8):a016972. https://doi.org/10.1101/cshperspect.a016972
El-Manzalawy Y, Dobbs D, Honavar V (2008) Predicting linear B-cell epitopes using string kernels. J Mol Recognit 21(4):243–255. https://doi.org/10.1002/jmr.893
Esmailnia E, Amani J, Gargari SLM (2020) Identification of novel vaccine candidate against Salmonella enterica serovar Typhi by reverse vaccinology method and evaluation of its immunization. Genomics 112(5):3374–3381. https://doi.org/10.1016/j.ygeno.2020.06.022
Flower D, Doytchinova I, Zaharieva N, Dimitrov I (2017) Immunogenicity prediction by VaxiJen: a ten year overview. J Proteom Bioinform 10:298–310. https://doi.org/10.4172/jpb.1000454
Frenck RW Jr, Baqar S, Alexander W, Dickey M, McNeal M, El-Khorazaty J, Baughman H, Hoeper A, Barnoy S, Suvarnapunya AE, Kaminski RW, Venkatesan MM (2018) A phase I trial to evaluate the safety and immunogenicity of WRSs2 and WRSs3; two live oral candidate vaccines against Shigella sonnei. Vaccine 36(32 Pt B):4880–4889. https://doi.org/10.1016/j.vaccine.2018.06.063
Garg A, Gupta D (2008) VirulentPred: a SVM based prediction method for virulent proteins in bacterial pathogens. BMC Bioinformatics 9(1):62. https://doi.org/10.1186/1471-2105-9-62
Gasteiger E, Hoogland C, Gattiker A, Se D, Wilkins MR, Appel RD, Bairoch A (2005) Protein identification and analysis tools on the ExPASy server. In: Walker JM (ed) The proteomics protocols handbook. Humana Press, Totowa, pp 571–607
Gilavand F, Marzban A, Ebrahimipour G, Soleimani N, Goudarzi M (2020) Designation of chitosan nano-vaccine based on MxiH antigen of Shigella flexneri with increased immunization capacity. Carbohydr Polym 232:115813. https://doi.org/10.1016/j.carbpol.2019.115813
Gregory M, Kaminski RW, Lugo-Roman LA, Galvez Carrillo H, Tilley DH, Baldeviano C, Simons MP, Reynolds ND, Ranallo RT, Suvarnapunya AE, Venkatesan MM, Oaks EV (2014) Development of an Aotus nancymaae model for Shigella vaccine immunogenicity and efficacy studies. Infect Immun 82(5):2027–2036. https://doi.org/10.1128/IAI.01665-13
Hattotuwagama CK, Guan P, Doytchinova IA, Zygouri C, Flower DR (2004) Quantitative online prediction of peptide binding to the major histocompatibility complex. J Mol Graph Model 22(3):195–207. https://doi.org/10.1016/s1093-3263(03)00160-8
Hebditch M, Carballo-Amador MA, Charonis S, Curtis R, Warwicker J (2017) Protein–Sol: a web tool for predicting protein solubility from sequence. Bioinformatics 33(19):3098–3100. https://doi.org/10.1093/bioinformatics/btx345
Jennison AV, Verma NK (2004) Shigella flexneri infection: pathogenesis and vaccine development. FEMS Microbiol Rev 28(1):43–58. https://doi.org/10.1016/j.femsre.2003.07.002
Jennison AV, Raqib R, Verma NK (2006) Immunoproteome analysis of soluble and membrane proteins of Shigella flexneri 2457T. World J Gastroenterol 12(41):6683–6688. https://doi.org/10.3748/wjg.v12.i41.6683
Kanehisa M, Goto S, Kawashima S, Nakaya A (2002) The KEGG databases at GenomeNet. Nucleic Acids Res 30(1):42–46. https://doi.org/10.1093/nar/30.1.42
Khare S, Alali W, Zhang S, Hunter D, Pugh R, Fang FC, Libby SJ, Adams LG (2010) Vaccination with attenuated Salmonella enterica Dublin expressing E coli O157:H7 outer membrane protein Intimin induces transient reduction of fecal shedding of E coli O157:H7 in cattle. BMC Vet Res 6:35. https://doi.org/10.1186/1746-6148-6-35
Kim JO, Rho S, Kim SH, Kim H, Song HJ, Kim EJ, Kim RY, Kim EH, Sinha A, Dey A, Yang JS, Song MK, Nandy RK, Czerkinsky C, Kim DW (2015) Shigella outer membrane protein PSSP-1 is broadly protective against Shigella infection. Clin Vaccine Immunol 22(4):381–388. https://doi.org/10.1128/cvi.00661-14
Kotloff KL, Nataro JP, Blackwelder WC, Nasrin D, Farag TH, Panchalingam S, Wu Y, Sow SO, Sur D, Breiman RF, Faruque AS, Zaidi AK, Saha D, Alonso PL, Tamboura B, Sanogo D, Onwuchekwa U, Manna B, Ramamurthy T, Kanungo S, Ochieng JB, Omore R, Oundo JO, Hossain A, Das SK, Ahmed S, Qureshi S, Quadri F, Adegbola RA, Antonio M, Hossain MJ, Akinsola A, Mandomando I, Nhampossa T, Acacio S, Biswas K, O'Reilly CE, Mintz ED, Berkeley LY, Muhsen K, Sommerfelt H, Robins-Browne RM, Levine MM (2013) Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet 382(9888):209–222. https://doi.org/10.1016/s0140-6736(13)60844-2
Krogh A, Larsson B, von Heijne G, Sonnhammer EL (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305(3):567–580. https://doi.org/10.1006/jmbi.2000.4315
Larsen MV, Lundegaard C, Lamberth K, Buus S, Lund O, Nielsen M (2007) Large-scale validation of methods for cytotoxic T-lymphocyte epitope prediction. BMC Bioinformatics 8:424. https://doi.org/10.1186/1471-2105-8-424
Leow CY, Kazi A, Hisyam Ismail CMK, Chuah C, Lim BH, Leow CH, Banga Singh KK (2020) Reverse vaccinology approach for the identification and characterization of outer membrane proteins of Shigella flexneri as potential cellular- and antibody-dependent vaccine candidates. Clin Exp Vaccine Res 9(1):15–25. https://doi.org/10.7774/cevr.2020.9.1.15
Liao Y, Deng J, Zhang A, Zhou M, Hu Y, Chen H, Jin M (2009) Immunoproteomic analysis of outer membrane proteins and extracellular proteins of Actinobacillus pleuropneumoniae JL03 serotype 3. BMC Microbiol 9(1):172. https://doi.org/10.1186/1471-2180-9-172
Ma ST, Ding GJ, Huang XW, Wang ZW, Wang L, Yu ML, Shi W, Jiang YP, Tang LJ, Xu YG, Li YJ (2018) Immunogenicity in chickens with orally administered recombinant chicken-borne Lactobacillus saerimneri expressing FimA and OmpC antigen of O78 avian pathogenic Escherichia coli. J Med Microbiol 67(3):441–451. https://doi.org/10.1099/jmm.0.000679
Madampage CA, Rawlyk N, Crockford G, Wang Y, White AP, Brownlie R, Van Donkersgoed J, Dorin C, Potter A (2015) Reverse vaccinology as an approach for developing Histophilus somni vaccine candidates. Biologicals 43(6):444–451. https://doi.org/10.1016/j.biologicals.2015.09.001
Maggiore L, Yu L, Omasits U, Rossi O, Dougan G, Thomson NR, Saul A, Choudhary JS, Gerke C (2016) Quantitative proteomic analysis of Shigella flexneri and Shigella sonnei Generalized Modules for Membrane Antigens (GMMA) reveals highly pure preparations. Int J Med Microbiol 306(2):99–108. https://doi.org/10.1016/j.ijmm.2015.12.003
Magnan C, Zeller M, Kayala M, Vigil A, Randall A, Felgner P, Baldi P (2010) High-throughput prediction of protein antigenicity using protein microarray data. Bioinformatics (Oxford, England) 26:2936–2943. https://doi.org/10.1093/bioinformatics/btq551
Mak TW, Saunders ME (2006) Immunity to pathogens. The immune response. Academic Press, Burlington, pp 641–694. https://doi.org/10.1016/B978-012088451-3.50024-7
Marteyn BS (2016) Shigella vaccine development: the model matters. JSM Trop Med Res 1(2):1011
Mattock E, Blocker AJ (2017) How do the virulence factors of Shigella work together to cause disease? Front Cell Infect Microbiol 7(64). https://doi.org/10.3389/fcimb.2017.00064
Medeiros PHQS, Bolick DT, Ledwaba SE, Kolling GL, Costa DVS, Oriá RB, Lima AÂM, Barry EM, Guerrant RL (2020) A bivalent vaccine confers immunogenicity and protection against Shigella flexneri and enterotoxigenic Escherichia coli infections in mice. NPJ Vaccines 5(1):30. https://doi.org/10.1038/s41541-020-0180-y
Meunier M, Guyard-Nicodème M, Hirchaud E, Parra A, Chemaly M, Dory D (2016) Identification of novel vaccine candidates against Campylobacter through reverse vaccinology. J Immunol Res 2016:5715790. https://doi.org/10.1155/2016/5715790
Mora M, Veggi D, Santini L, Pizza M, Rappuoli R (2003) Reverse vaccinology. Drug Discov Today 8(10):459–464. https://doi.org/10.1016/s1359-6446(03)02689-8
Morris CR, Grassel CL, Redman JC, Sahl JW, Barry EM, Rasko DA (2013) Characterization of intracellular growth regulator icgR by utilizing transcriptomics to identify mediators of pathogenesis in Shigella flexneri. Infect Immun 81(9):3068–3076. https://doi.org/10.1128/iai.00537-13
Naz A, Awan FM, Obaid A, Muhammad SA, Paracha RZ, Ahmad J, Ali A (2015) Identification of putative vaccine candidates against Helicobacter pylori exploiting exoproteome and secretome: a reverse vaccinology based approach. Infect Genet Evol 32:280–291. https://doi.org/10.1016/j.meegid.2015.03.027
Okafor AACN (2018) Shigella (Shigellosis). StatPearls [Internet] Beth Israel Deaconess Medical Center Creighton University. NBK482337
Oprea M, Antohe F (2013) Reverse-vaccinology strategy for designing T-cell epitope candidates for Staphylococcus aureus endocarditis vaccine. Biologicals 41:148–153
Pore D, Chakrabarti MK (2013) Outer membrane protein A (OmpA) from Shigella flexneri 2a: a promising subunit vaccine candidate. Vaccine 31(36):3644–3650. https://doi.org/10.1016/j.vaccine.2013.05.100
Pore D, Chakrabarti MK (2016) An approach to identify and characterize a subunit candidate Shigella vaccine antigen. Methods Mol Biol 1403:433–444. https://doi.org/10.1007/978-1-4939-3387-7_24
Psonis JJ, Thanassi DG (2019) Therapeutic approaches targeting the assembly and function of chaperone-usher pili. EcoSal Plus 8(2). https://doi.org/10.1128/ecosalplus.ESP-0033-2018
Rashid MI, Naz A, Ali A, Andleeb S (2017) Prediction of vaccine candidates against Pseudomonas aeruginosa: an integrated genomics and proteomics approach. Genomics 109(3–4):274–283. https://doi.org/10.1016/j.ygeno.2017.05.001
Riddle MS, Kaminski RW, Williams C, Porter C, Baqar S, Kordis A, Gilliland T, Lapa J, Coughlin M, Soltis C, Jones E, Saunders J, Keiser PB, Ranallo RT, GormLey R, Nelson M, Turbyfill KR, Tribble D, Oaks EV (2011) Safety and immunogenicity of an intranasal Shigella flexneri 2a Invaplex 50 vaccine. Vaccine 29(40):7009–7019. https://doi.org/10.1016/j.vaccine.2011.07.033
Saha S, Raghava GPS (2004) BcePred: Prediction of continuous B-cell epitopes in antigenic sequences using physico-chemical properties. In: Berlin, Heidelberg, 2004. Artificial Immune Systems, vol 3239. Springer, Berlin Heidelberg, pp 197–204. https://doi.org/10.1007/978-3-540-30220-9_16
Saha S, Raghava GPS (2006) Prediction of continuous B-cell epitopes in an antigen using recurrent neural network. Proteins: structure, function. Bioinformatics 65(1):40–48. https://doi.org/10.1002/prot.21078
Schussek S, Trieu A, Doolan DL (2014) Genome- and proteome-wide screening strategies for antigen discovery and immunogen design. Biotechnol Adv 32(2):403–414. https://doi.org/10.1016/j.biotechadv.2013.12.006
Sereny B (1955) Experimental shigella keratoconjunctivitis; a preliminary report. Acta Microbiol Acad Sci Hung 2(3):293–296
Sette A, Rappuoli R (2010) Reverse vaccinology: developing vaccines in the era of genomics. Immunity 33(4):530–541. https://doi.org/10.1016/j.immuni.2010.09.017
Sharma D, Yagnik B, Baksi R, Desai N, Padh H, Desai P (2017) Shigellosis murine model established by intraperitoneal and intranasal route of administration: a comparative comprehension overview. Microbes Infect 19(1):47–54. https://doi.org/10.1016/j.micinf.2016.09.002
Smialowski P, Doose G, Torkler P, Kaufmann S, Frishman D (2012) PROSO II – a new method for protein solubility prediction. FEBS J 279(12):2192–2200. https://doi.org/10.1111/j.1742-4658.2012.08603.x
Son WG, Graham TA, Gannon VP (2002) Immunological characterization of Escherichia coli O157:H7 intimin gamma1. Clin Diagn Lab Immunol 9(1):46–53. https://doi.org/10.1128/cdli.9.1.46-53.2002
Studier FW (2005) Protein production by auto-induction in high density shaking cultures. Protein Expr Purif 41(1):207–234. https://doi.org/10.1016/j.pep.2005.01.016
Talukdar S, Zutshi S, Prashanth KS, Saikia KK, Kumar P (2014) Identification of potential vaccine candidates against Streptococcus pneumoniae by reverse vaccinology approach. Appl Biochem Biotechnol 172(6):3026–3041. https://doi.org/10.1007/s12010-014-0749-x
Tusnády GE, Simon I (2001) The HMMTOP transmembrane topology prediction server. Bioinformatics 17(9):849–850. https://doi.org/10.1093/bioinformatics/17.9.849
Vishnu US, Sankarasubramanian J, Gunasekaran P, Rajendhran J (2017) Identification of potential antigens from non-classically secreted proteins and designing novel multitope peptide vaccine candidate against Brucella melitensis through reverse vaccinology and immunoinformatics approach. Infect Genet Evol 55:151–158. https://doi.org/10.1016/j.meegid.2017.09.015
Vita R, Mahajan S, Overton JA, Dhanda SK, Martini S, Cantrell JR, Wheeler DK, Sette A, Peters B (2019) The Immune Epitope Database (IEDB): 2018 update. Nucleic Acids Res 47(D1):D339–d343. https://doi.org/10.1093/nar/gky1006
Wang L, Zhu Z, Qian H, Li Y, Chen Y, Ma P, Gu B (2019) Comparative genome analysis of 15 clinical Shigella flexneri strains regarding virulence and antibiotic resistance. AIMS Microbiol 5(3):205–222. https://doi.org/10.3934/microbiol.2019.3.205
Williams PCM, Berkley JA (2018) Guidelines for the treatment of dysentery (shigellosis): a systematic review of the evidence. Paediatr Int Child Health 38(sup1):S50–s65. https://doi.org/10.1080/20469047.2017.1409454
Yang JY, Lee SN, Chang SY, Ko HJ, Ryu S, Kweon MN (2014) A mouse model of shigellosis by intraperitoneal infection. J Infect Dis 209(2):203–215. https://doi.org/10.1093/infdis/jit399
Ying T, Wang H, Li M, Wang J, Wang J, Shi Z, Feng E, Liu X, Su G, Wei K, Zhang X, Huang P, Huang L (2005) Immunoproteomics of outer membrane proteins and extracellular proteins of Shigella flexneri 2a 2457T. Proteomics 5(18):4777–4793. https://doi.org/10.1002/pmic.200401326
Yu CS, Chen YC, Lu CH, Hwang JK (2006) Prediction of protein subcellular localization. Proteins 64(3):643–651. https://doi.org/10.1002/prot.21018
Yu NY, Wagner JR, Laird MR, Melli G, Rey S, Lo R, Dao P, Sahinalp SC, Ester M, Foster LJ, Brinkman FSL (2010) PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26(13):1608–1615. https://doi.org/10.1093/bioinformatics/btq249
Acknowledgments
This study was performed in the Department of Cell Biology, Shahed University. The authors are grateful for the contributions and financial supports of the research deputy throughout this study. The authors also thank Dr. Shakiba Darvish Alipour for her technical assistance in this work.
Funding
This work was funded by the deputy research (grant no: 957586003), Shahed University, Tehran, Iran.
Author information
Authors and Affiliations
Contributions
LM designed the project and conceptualized the study. All the in silico studies and statistical analyses were performed by AH and supervised by JA. Also, AH carried out laboratory experiments and drafted the manuscript. LM took the lead role in manuscript preparation.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All the animal-related experiments were conducted in accordance with ethical standards and with animal care guidelines confirmed by the Animal Care and Ethical Committee of Shahed University. All applicable international guidelines for the care and use of animals were followed (Ethical Code: 957586003). This article does not contain any studies with human participants performed by any of the authors.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
ESM 1
(XLSX 75.3 kb)
Rights and permissions
About this article
Cite this article
Hajialibeigi, A., Amani, J. & Gargari, S.L.M. Identification and evaluation of novel vaccine candidates against Shigella flexneri through reverse vaccinology approach. Appl Microbiol Biotechnol 105, 1159–1173 (2021). https://doi.org/10.1007/s00253-020-11054-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-020-11054-4