Abstract
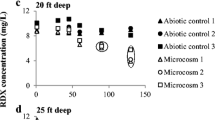
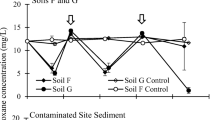
1,4-Dioxane, a probable human carcinogen, is a co-contaminant at many chlorinated solvent-contaminated sites. Although numerous 1,4-dioxane-degrading aerobic bacteria have been isolated, almost no information exists on the microorganisms able to degrade this chemical under anaerobic conditions. Here, the potential for 1,4-dioxane biodegradation was examined using multiple inocula and electron acceptor amendments. The inocula included uncontaminated agricultural soils and river sediments as well as sediments from two 1,4-dioxane contaminated sites. Five separate experiments involved the examination of triplicate live microcosms and abiotic controls for approximately 1 year. Compound-specific isotope analysis (CSIA) was used to further investigate biodegradation in a subset of the microcosms. Also, DNA was extracted from microcosms exhibiting 1,4-dioxane biodegradation for microbial community analysis using 16S rRNA gene amplicon high-throughput sequencing. Given the long incubation periods, it is likely that electron acceptor depletion occurred and methanogenic conditions eventually dominated. The iron/EDTA/humic acid or sulfate amendments did not result in 1,4-dioxane biodegradation in the majority of cases. 1,4-dioxane biodegradation was most commonly observed in the nitrate amended and no electron acceptor treatments. Notably, both contaminated site sediments illustrated removal in the samples compared to the abiotic controls in the no electron acceptor treatment. However, it is important to note that the degradation was slow (with concentration reductions occurring over approximately 1 year). In two of the three cases examined, CSIA provided additional evidence for 1,4-dioxane biodegradation. In one case, the reduction in 1,4-dioxane in the samples comparing the controls was likely too low for the method to detect a significant 13C/12C enrichment. Further research is required to determine the value of measuring 2H/1H for generating evidence for the biodegradation of this chemical. The microbial community analysis indicated that the phylotypes unclassified Comamonadaceae and 3 genus incertae sedis were more abundant in 1,4-dioxane-degrading microcosms compared to the live controls (no 1,4-dioxane) in microcosms inoculated with contaminated and uncontaminated sediment, respectively. The relative abundance of known 1,4-dioxane degraders was also investigated at the genus level. The soil microcosms were dominated primarily by Rhodanobacter with lower relative abundance values for Pseudomonas, Mycobacterium, and Acinetobacter. The sediment communities were dominated by Pseudomonas and Rhodanobacter. Overall, the current study indicates 1,4-dioxane biodegradation under anaerobic and, likely methanogenic conditions, is feasible. Therefore, natural attenuation may be an appropriate cleanup technology at sites where time is not a limitation.









Similar content being viewed by others
References
Adams CD, Scanlan PA, Secrist ND (1994) Oxidation and biodegradability enhancement of 1,4-dioxane using hydrogen peroxide and ozone. Environ Sci Technol 28(11):1812–1818
Adamson DT, Mahendra S, Walker KL, Rauch SR, Sengupta S, Newell CJ (2014) A multisite survey to identify the scale of the 1,4-dioxane problem at contaminated groundwater sites. Environ Sci Tech Let 1(5):254–258
Adamson DT, Anderson RH, Mahendra S, Newell CJ (2015) Evidence of 1,4-dioxane attenuation at groundwater sites contaminated with chlorinated solvents and 1,4-dioxane. Environ Sci Technol 49(11):6510–6518. https://doi.org/10.1021/acs.est.5b00964
Bennett P, Hyman M, Smith C, El Mugammar H, Chu MY, Nickelsen M, Aravena R (2018) Enrichment with carbon-13 and deuterium during monooxygenase-mediated biodegradation of 1,4-dioxane. Environ Sci Tech Let 5(3):148–153. https://doi.org/10.1021/acs.estlett.7b00565
Bernhardt D, Diekmann H (1991) Degradation of dioxane, tetrahydrofuran and other cyclic ethers by an environmental Rhodococcus strain. Appl Microbiol Biot 36(1):120–123
Bhushan B, Halasz A, Hawari J (2006) Effect of iron(III), humic acids and anthraquinone-2,6-disulfonate on biodegradation of cyclic nitramines by Clostridium sp. EDB2. J Appl Microbiol 100(3):555–563. https://doi.org/10.1111/j.1365-2672.2005.02819.x
Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R (2011) Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. P Natl Acad Sci USA 108:4516–4522
Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, Gormley N, Gilbert JA, Smith G, Knight R (2012) Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 6(8):1621–1624
Chen DZ, Jin XJ, Chen J, Ye JX, Jiang NX, Chen JM (2016) Intermediates and substrate interaction of 1,4-dioxane degradation by the effective metabolizer Xanthobacter flavus DT8. Int Biodeterior Biodegradation 106:133–140. https://doi.org/10.1016/j.ibiod.2015.09.018
Chu MYJ, Bennett P, Dolan M, Hyman M, Anderson R, Bodour A, Peacock A (2009) Using aerobic cometabolic biodegradation and groundwater recirculation to treat 1,4-dioxane and co-contaminants in a dilute plume. Paper presented at the tenth international conference on the remediation of chlorinated and recalcitrant compounds. Baltimore, Maryland
Cupples AM (2008) Real-time PCR quantification of Dehalococcoides populations: Methods and applications. J Microbiol Methods 72(1):1–11. https://doi.org/10.1016/j.mimet.2007.11.005
Cupples AM, Spormann AM, McCarty PL (2003) Growth of a Dehalococcoides-like microorganism on vinyl chloride and cis-dichloroethene as electron acceptors as determined by competitive PCR. Appl Environ Microbiol 69(2):953–959
Deng DY, Li F, Wu C, Li MY (2018) Synchronic biotransformation of 1,4-dioxane and 1,1-dichloroethylene by a gram-negative propanotroph Azoarcus sp DD4. Environ Sci Tech Let 5(8):526. https://doi.org/10.1021/acs.estlett.8b00312
DeRosa CT, Wilbur S, Holler J, Richter P, Stevens YW (1996) Health evaluation of 1,4-dioxane. Toxicol & Indust Health 12(1):1–43
Freedman DL, Gossett JM (1989) Biological reductive dechlorination of tetrachloroethylene and trichloroethylene to ethylene under methanogenic conditions. Appl Environ Microbiol 55(9):2144–2151
Gedalanga P, Madison A, Miao Y, Richards T, Hatton J, DiGuiseppi WH, Wilson J, Mahendra S (2016) A multiple lines of evidence framework to evaluate intrinsic biodegradation of 1,4-dioxane. Remediation 27(1):93–114. https://doi.org/10.1002/rem.21499
Hand S, Wang BX, Chu KH (2015) Biodegradation of 1,4-dioxane: effects of enzyme inducers and trichloroethylene. Sci Total Environ 520:154–159
He J, Ritalahti KM, Yang K-L, Koeningsberg SS, Löffler FE (2003) Detoxification of vinyl chloride to ethene coupled to growth of an anaerobic bacterium. Nature 424:62–65
Holliger C, Wohlfarth G, Diekert G (1998) Reductive dechlorination in the energy metabolism of anaerobic bacteria. FEMS Microbiol Rev 22(5):383–398
Huang HL, Shen DS, Li N, Shan D, Shentu JL, Zhou YY (2014) Biodegradation of 1,4-dioxane by a novel strain and its biodegradation pathway. Water Air Soil Poll 225:9
Hunkeler D, Meckenstock RU, Lollar BS, Schmidt TC, Wilson JT (2008) A guide for assessing biodegradation and source identification of organic ground water contaminants using compound specific isotope analysis (CSIA). USEPA, Oklahoma
Inoue D, Tsunoda T, Sawada K, Yamamoto N, Saito Y, Sei K, Ike M (2016) 1,4-Dioxane degradation potential of members of the genera Pseudonocardia and Rhodococcus. Biodegrad 27(4–6):277–286. https://doi.org/10.1007/s10532-016-9772-7
Inoue D, Tsunoda T, Yamamoto N, Ike M, Sei K (2018) 1,4-Dioxane degradation characteristics of Rhodococcus aetherivorans JCM 14343. Biodegrad 29(3):301–310. https://doi.org/10.1007/s10532-018-9832-2
Isaka K, Udagawa M, Sei K, Ike M (2016) Pilot test of biological removal of 1,4-dioxane from a chemical factory wastewater by gel carrier entrapping Afipia sp strain D1. J Hazard Mater 304:251–258. https://doi.org/10.1016/j.jhazmat.2015.10.066
Kampfer P, Kroppenstedt RM (2004) Pseudonocardia benzenivorans sp nov. Int J Syst Evol Micr 54:749–751. https://doi.org/10.1099/ijs.0.02825-0
Kim YM, Jeon JR, Murugesan K, Kim EJ, Chang YS (2009) Biodegradation of 1,4-dioxane and transformation of related cyclic compounds by a newly isolated Mycobacterium sp PH-06. Biodegrad 20(4):511–519
Kohlweyer U, Thiemer B, Schrader T, Andreesen JR (2000) Tetrahydrofuran degradation by a newly isolated culture of Pseudonocardia sp strain K1. Fems Microbiol Lett 186(2):301–306. https://doi.org/10.1111/j.1574-6968.2000.tb09121.x
Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD (2013) Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microb 79(17):5112–5120. https://doi.org/10.1128/Aem.01043-13
Lan RS, Smith CA, Hyman MR (2013) Oxidation of cyclic ethers by alkane-grown Mycobacterium vaccae JOB5. Remediation 23(4):23–42. https://doi.org/10.1002/rem.21364
Li MY, Fiorenza S, Chatham JR, Mahendra S, Alvarez PJJ (2010) 1,4-Dioxane biodegradation at low temperatures in Arctic groundwater samples. Water Res 44(9):2894–2900. https://doi.org/10.1016/j.watres.2010.02.007
Lippincott D, Streger SH, Schaefer CE, Hinkle J, Stormo J, Steffan RJ (2015) Bioaugmentation and propane biosparging for in situ biodegradation of 1,4-dioxane. Ground Water Monit R 35(2):81–92
Löffler FE, Yan J, Ritalahti KM, Adrian L, Edwards EA, Konstantinidis KT, Müller JA, Fullerton H, Zinder SH, Spormann AM (2013) Dehalococcoides mccartyi gen. Nov., sp. nov., obligately organohalide-respiring anaerobic bacteria relevant to halogen cycling and bioremediation, belong to a novel bacterial class, Dehalococcoidia classis nov., order Dehalococcoidales Ord. Nov. and family Dehalococcoidaceae fam. Nov., within the phylum Chloroflexi. Int J Syst Evol Microbiol 63(Pt 2):625–635
Lu ZD, Sun WJ, Li C, Ao XW, Yang C, Li SM (2019) Bioremoval of non-steroidal anti-inflammatory drugs by Pseudoxanthomonas sp. DIN-3 isolated from biological activated carbon process. Water Res 161:459–472. https://doi.org/10.1016/j.watres.2019.05.065
Mahendra S, Alvarez-Cohen L (2005) Pseudonocardia dioxanivorans sp nov., a novel actinomycete that grows on 1,4-dioxane. Int J Syst Evol Micr 55:593–598. https://doi.org/10.1099/ijs.0.63085-0
Mahendra S, Alvarez-Cohen L (2006) Kinetics of 1,4-dioxane biodegradation by monooxygenase-expressing bacteria. Environ Sci Technol 40(17):5435–5442. https://doi.org/10.1021/es060714v
Major DW, McMaster ML, Cox EE, Edwards EA, Dworatzek SM, Hendrickson ER, Starr MG, Payne JA, Buonamici LW (2002) Field demonstration of successful bioaugmentation to achieve dechlorination of tetrachloroethene to ethene. Environ Sci Technol 36(23):5106–5116
Masuda H (2009) Identification and characterization of monooxygenase enzymes involved in 1,4-dioxane degradation in Pseudonocardia sp. strain ENV478, Mycobacterium sp. strain ENV421, and Nocardia sp. strain ENV425. Rutgers the State University of new Jersey and University of Medicine and dentistry of New Jersey
Matsui R, Takagi K, Sakakibara F, Abe T, Shiiba K (2016) Identification and characterization of 1,4-dioxane-degrading microbe separated from surface seawater by the seawater-charcoal perfusion apparatus. Biodegrad 27(2–3):155–163. https://doi.org/10.1007/s10532-016-9763-8
Maymó-Gatell X, Chien Y-T, Gossett JM, Zinder SH (1997) Isolation of a bacterium that reductively dechlorinates tetrachloroethene to ethene. Science 276(6):1568–1571
Mohr T, Stickney J, DiGuiseppi W (2010) Environmental investigation and remediation: 1,4-Dioxane and other solvent stabilizers. CRC Press, Boca Raton
Naumoff DG, Dedysh SN (2018) Bacteria from poorly studied phyla as a potential source of new enzymes: beta-galactosidases from Planctomycetes and Verrucomicrobia. Microbiology 87(6):796–805. https://doi.org/10.1134/S0026261718060127
Parales RE, Adamus JE, White N, May HD (1994) Degradation of 1,4-dioxane by an Actinomycete in pure culture. Appl Environ Microb 60(12):4527–4530
Parks DH, Tyson GW, Hugenholtz P, Beiko RG (2014) STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics 30(21):3123–3124
Pooley KE, Blessing M, Schmidt TC, Haderlein SB, MacQuarrie KTB, Prommer H (2009) Aerobic biodegradation of chlorinated ethenes in a fractured bedrock aquifer: quantitative assessment by compound-specific isotope analysis (CSIA) and reactive transport modeling. Environ Sci Technol 43(19):7458–7464. https://doi.org/10.1021/es900658n
Pornwongthong P (2014) Stable isotopic and molecular biological tools to validate biodegradation of 1,4-dioxane. University of California, Los Angeles
Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Gloeckner FO (2007) SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nuc Acids Res 35(21):7188–7196. https://doi.org/10.1093/nar/gkm864
Pugazhendi A, Banu JR, Dhavamani J, Yeom IT (2015) Biodegradation of 1,4-dioxane by Rhodanobacter AYS5 and the role of additional substrates. Ann Microbiol 65(4):2201–2208. https://doi.org/10.1007/s13213-015-1060-y
Qian Y, Chen K, Liu Y, Li J (2019) Assessment of hexachlorcyclohexane biodegradation in contaminated soil by compound-specific stable isotope analysis. Environ Pollut 254:113008
Rodriguez FJB (2016) Evaluation of 1,4-dioxane biodegradation under aerobic and anaerobic conditions. Clemson University
Rosell M, Häggblom MM, Richnow H-H (2007) Compound-specific isotope analysis (CSIA) to characterise degradation pathways and to quantify in-situ degradation of fuel oxygenates and other fuel-derived contaminants. In: Barceló D (ed) Fuel Oxygenates. Springer, Berlin Heidelberg, Berlin, Heidelberg, pp 99–119
Schloss PD (2009) A high-throughput DNA sequence aligner for microbial ecology studies. PLoS One 4(12). https://doi.org/10.1371/journal.pone.0008230
Schloss PD (2013) MiSeq SOP. PUblisher. http://www.mothur.org/wiki/MiSeq_SOP Accessed 15th Sept 2013
Sei K, Miyagaki K, Kakinoki T, Fukugasako K, Inoue D, Ike M (2013a) Isolation and characterization of bacterial strains that have high ability to degrade 1,4-dioxane as a sole carbon and energy source. Biodegrad 24(5):665–674
Sei K, Oyama M, Kakinoki T, Inoue D, Ike M (2013b) Isolation and characterization of tetrahydrofuran- degrading bacteria for 1,4-dioxane-containing wastewater treatment by co-metabolic degradation. J Water Environ Technol 11:11–19. https://doi.org/10.2965/jwet.2013.11
Shen W, Chen H, Pan S (2008) Anaerobic biodegradation of 1,4-dioxane by sludge enriched with iron-reducing microorganisms. Bioresour Technol 99(7):2483–2487. https://doi.org/10.1016/j.biortech.2007.04.054
Stefan MI, Bolton JR (1998) Mechanism of the degradation of 1,4-dioxane in dilute aqueous solution using the UV hydrogen peroxide process. Environ Sci Technol 32(11):1588–1595
Steffan RJ (2007) ER-1422: biodegradation of 1,4-dioxane. vol Final Report. SERDP and ESTCP
Steffan RJ, Vainberg S (2013) Production and handling of Dehalococcodies bioaugmentation cultures. In: Stroo HF, Leeson A, Ward CW (eds) Bioaugmentation for groundwater remediation. SERDP and ESTCP Remediation Technology. Springer, New York
Steffan RJ, McClay K, Vainberg S, Condee CW, Zhang DL (1997) Biodegradation of the gasoline oxygenates methyl tert-butyl ether, ethyl tert-butyl ether, and tert-amyl methyl ether by propane-oxidizing bacteria. Appl Environ Microb 63(11):4216–4222
Steffan RJ, McClay KR, Masuda H, Zylstra GJ (2007) Biodegradatdion of 1,4-dioxane. Strategic environmental Research and Development program: ER-1422 final report
Stringfellow WT, Alvarez-Cohen L (1999) Evaluating the relationship between the sorption of PAHs to bacterial biomass and biodegradation. Water res 33(11):2535–2544. https://doi.org/10.1016/S0043-1354(98)00497-7
Sun BZ, Ko K, Ramsay JA (2011) Biodegradation of 1,4-dioxane by a Flavobacterium. Biodegrad 22(3):651–659. https://doi.org/10.1007/s10532-010-9438-9
Sung Y, Ritalahti KM, Apkarian RP, Loffler FE (2006) Quantitative PCR confirms purity of strain GT, a novel trichloroethene-to-ethene-respiring Dehalococcoides isolate. Appl Environ Microbiol 72(3):1980–1987
Tanaka Y, Matsuzawa H, Tamaki H, Tagawa M, Toyama T, Kamagata Y, Mori K (2017) Isolation of novel bacteria including rarely cultivated phyla, Acidobacteria and Verrucomicrobia, from the roots of emergent plants by simple culturing method. Microbes Environ 32(3):288–292. https://doi.org/10.1264/jsme2.ME17027
Vainberg S, McClay K, Masuda H, Root D, Condee C, Zylstra GJ, Steffan RJ (2006) Biodegradation of ether pollutants by Pseudonocardia sp strain ENV478. Appl Environ Microb 72(8):5218–5224
Vainberg S, Condee CW, Steffan RJ (2009) Large-scale production of bacterial consortia for remediation of chlorinated solvent-contaminated groundwater. J Ind Microbiol Biot 36(9):1189–1197. https://doi.org/10.1007/S10295-009-0600-5
Wang Y, Huang Y, Huckins JN, Petty JD (2004) Compound-specific carbon and hydrogen isotope analysis of sub-parts per billion level waterborne petroleum hydrocarbons. Environ Sci Technol 38(13):3689–3697. https://doi.org/10.1021/es035470i
Whittenbury R, Phillips KC, Jf W (1970) Enrichment, isolation and some properties of methane-utilizing bacteria. J Gen Microbiol 61:–205, 18. https://doi.org/10.1099/00221287-61-2-205
Willems A (2014) The Family Comamonadaceae. In: Eugene Rosenberg (Editor-in-Chief) EFD, Stephen Lory, Erko Stackebrandt and Fabiano Thompson (Eds.) (ed) The prokaryotes: Alphaproteobacteria and betaproteobacteria. 4th edn. Springer-Verlag Berlin Heidelberg
Yamamoto N, Saito Y, Inoue D, Sei K, Ike M (2018) Characterization of newly isolated Pseudonocardia sp N23 with high 1,4-dioxane-degrading ability. J Biosci Bioeng 125(5):552–558. https://doi.org/10.1016/j.jbiosc.2017.12.005
Zenker MJ, Borden RC, Barlaz MA (2003) Occurrence and treatment of 1,4-dioxane in aqueous environments. Env Eng Sci 20(5):423–432
Acknowledgments
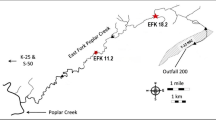
We would like to thank Dr. Dan Jones and Dr. Scott Smith at the Mass Spectrometry Laboratory at the Research Technology Support Facility (MSU) for 1,4-dioxane analytical method support and also to Dr. Anthony Danko (Naval Facilities Engineering Command) for providing the contaminated site sediments.
Funding
Support for this research was also provided by the NSF Long-term Ecological Research Program (DEB 1832042) at the Kellogg Biological Station and by Michigan State University AgBioResearch.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors declare no conflict of interest.
Ethical statement
This work is supported by a grant awarded to Dr. Cupples from Strategic Environmental Research and Development Program (SERDP). Contract Number: W912HQ-17-C-0006.
Human and animal rights and informed consent
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(PDF 30 kb)
Rights and permissions
About this article
Cite this article
Ramalingam, V., Cupples, A.M. Anaerobic 1,4-dioxane biodegradation and microbial community analysis in microcosms inoculated with soils or sediments and different electron acceptors. Appl Microbiol Biotechnol 104, 4155–4170 (2020). https://doi.org/10.1007/s00253-020-10512-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-020-10512-3