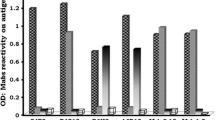
Abstract
We developed a fast, rabies virus-free, in vitro method, based on a blocking ELISA (bELISA), to detect and accurately quantify anti-rabies glycoprotein antibodies in serum of several animal species. In this method, purified rabies virus-like particles (VLPs) are used as antigen to coat the plates, while the presence of specific rabies immunoglobulins is revealed through blocking the recognition of these VLPs by a biotinylated monoclonal antibody. A quality by design approach was carried out in order to optimize the method performance, improving the sensitivity and, thereby, reducing the limit of detection of this assay. After the method validation, we confirmed that the bELISA method is able to detect a concentration of 0.06 IU/mL rabies immunoglobulins, titer lower than the 0.5 IU/mL cutoff value established as indication for correct vaccination. Further, we assessed the correlation between bELISA, the MNT, and the Platelia methods, confirming the accuracy of this new assay. On the other hand, precision was evaluated, obtaining acceptable repeatability and intermediate precision values, showing that this bELISA could be proposed as a potential alternative method, replacing the gold standard techniques in vaccination schemes and becoming a routine control technique within regional rabies surveillance programs.





Similar content being viewed by others
References
Briggs DJ, Schweitzer K (1997) Importation of dogs and cats to rabies-free areas of the world. Vet Clin North Am Small Anim Pract 31:573–583. https://doi.org/10.1016/S0195-5616(01)50610-5
Celis E, Ou D, Dietzschold B, Koprowski H (1988) Recognition of rabies and rabies-related viruses by T cells derived from human vaccine recipients. J Virol 62:3128–3134
Cliquet F, Aubert M, Sag L (1998) Development of a fluorescent antibody virus neutralisation test (FAVN test) for the quantitation of rabies-neutralising antibody. 212:79–87. https://doi.org/10.1016/s0022-1759(97)00212-3
Cliquet F, McElhinney LM, Servat A, Boucher JM, Lowings JP, Goddard T, Mansfield KL, Fooks AR (2004) Development of a qualitative indirect ELISA for the measurement of rabies virus-specific antibodies from vaccinated dogs and cats. J Virol Methods 117:1–8. https://doi.org/10.1016/j.jviromet.2003.12.001
Cox JH, Dietzschold B, Schneider LG (1977) Rabies virus glycoprotein. II. Biological and serological characterization. Infect Immun 16:754–759
Derringer G, Suich R (1980) Simultaneous optimization of several response variables. J Qual Technol 12:214–219. https://doi.org/10.1080/00224065.1980.11980968
Ertl HCJ (2009) Novel vaccines to human rabies. PLoS Negl Trop Dis 3:e515. https://doi.org/10.1371/journal.pntd.0000515
FDA (2018) Bioanalytical method validation guidance for industry bioanalytical method validation guidance for industry. https://www.fda.gov/files/drugs/published/Bioanalytical-Method-Validation-Guidance-for-Industry.pdf
Fehlner-gardiner C, Wandeler AI (2015) Detection of rabies virus antibodies by competitive enzyme-linked immunosorbent assay, 2nd edn, Elsevier Inc
Ferreira SLC, Bruns RE, Ferreira HS, Matos GD, David JM, Brandao GC, da Silva E, Portugal L, dos Reis P, Souza A, dos Santos W (2007) Box-Behnken design: An alternative for the optimizacion of analytical methods. Anal Chim Acta 597:179–186 https://doi.org/10.1016/j.aca.2007.07.011
Feyssaguet M, Dacheux L, Audry L, Compoint A, Morize JL, Blanchard I, Bourhy H (2007) Multicenter comparative study of a new ELISA, PLATELIA RABIES II, for the detection and titration of anti-rabies glycoprotein antibodies and comparison with the rapid fluorescent focus inhibition test (RFFIT) on human samples from vaccinated and non-vacci. Vaccine 25:2244–2251. https://doi.org/10.1016/j.vaccine.2006.12.012
Fontana D, Kratje R, Etcheverrigaray M, Prieto C (2014) Rabies virus-like particles expressed in HEK293 cells. Vaccine. https://doi.org/10.1016/j.vaccine.2014.02.031
Fontana D, Kratje R, Etcheverrigaray M, Prieto C (2015) Immunogenic virus-like particles continuously expressed in mammalian cells as a veterinary rabies vaccine candidate. Vaccine. https://doi.org/10.1016/j.vaccine.2015.03.088
Fontana D, Marsili F, Garay E, Battagliotti J, Etcheverrigaray M, Kratje R, Prieto C (2019) A simplified roller bottle platform for the production of a new generation VLPs rabies vaccine for veterinary applications. Comp Immunol Microbiol Infect Dis 65:70–75. https://doi.org/10.1016/j.cimid.2019.04.009
Galhardo JA, de Azevedo CS, Remonti BR, Neres Gonçalves VM, Azevedo Marques NT, Borges LO, Ahad das Neves D (2019) Canine rabies in the Brazil-Bolivia border region from 2006 to 2014. Ann Glob Health 85:1–5. https://doi.org/10.5334/aogh.2334
ICH (2005) ICH. Validation of analytical procedures: text and methodology Q2 (R1). 1–13. https://www.ema.europa.eu/en/documents/scientific-guideline/ich-q-2-r1-validation-analytical-procedures-text-methodology-step-5_en.pdf
International Committee on Taxonomy of Viruses (2019) Online 10th Report, https://ictv.global/report/ Accesed 15 January 2020
Jáuregui G, Fontana D, Micheloud J, Prieto C, Delgado F (2019) Evaluation of new antibodies for the detection of rabies virus in formalin fixed brain tissue samples. Brazilian J Vet Pathol 12:27–32. https://doi.org/10.24070/bjvp.1983-0246.v12i2p27-32
Korimbocus J, Dehay N, Tordo N, Cano F, Morgeaux S (2016) Development and validation of a quantitative competitive ELISA for potency testing of equine anti rabies sera with other potential use. Vaccine 34:3310–3316. https://doi.org/10.1016/j.vaccine.2016.04.086
Laemmli, U (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685. https://doi.org/10.1038/227680a0
Leardi R (2009) Experimental design in chemistry: A tutorial. Anal Chim Acta 652(1):161–172. https://doi.org/10.1016/j.aca.2009.06.015
Moore SM, Hanlon CA (2010) Rabies-specific antibodies: measuring surrogates of protection against a fatal disease. PLoS Negl Trop Dis 4:e595. https://doi.org/10.1371/journal.pntd.0000595
Moore SM, Gordon CR, Hanlon CA (2013) Measures of rabies immunity. In: Jackson AC (ed) Rabies, 3rd edn. Elsevier, San Diego, pp 461–496
Nicholson KG, Prestage H (1982) Enzyme-linked immunosorbent assay: a rapid reproducible test for the measurement of rabies antibody. J Med Virol 9:43–49. https://doi.org/10.1002/jmv.1890090107
Ockuly RA, Weese ML, Smucker BJ, Edwards DJ, Chang L (2017) Response surface experiments: A meta-analysis. Chemom. Intell Lab Syst 154:64–75. https://doi.org/10.1016/j.chemolab.2017.03.009
Olivieri AC (2015) Practical guidelines for reporting results in single- and multi-component analytical calibration: a tutorial. Anal Chim Acta 868:10–22. https://doi.org/10.1016/j.aca.2015.01.017
Schnell MJ, McGettigan JP, Wirblich C, Papaneri A (2010) The cell biology of rabies virus: using stealth to reach the brain. Nat Rev Microbiol 8:51–61. https://doi.org/10.1038/nrmicro2260
Servat A, Feyssaguet M, Blanchard I, Morize JL, Schereffer JL, Boue F, Cliquet F (2007) A quantitative indirect ELISA to monitor the effectiveness of rabies vaccination in domestic and wild carnivores. J Immunol Methods 318:1–10. https://doi.org/10.1016/j.jim.2006.07.026
Smith JS, Yager PA, Baer GM (1973) A rapid reproducible test for determining rabies neutralizing antibody. Bull World Health Organ 48:535–541
Vanaja K, Rani RHS (2007) Design of experiments: concept and applications of plackett burman design. Res Regul Aff 24(1):1–23. https://doi.org/10.1080/10601330701220520
Webster LT, Dawson JR (1935) Early diagnosis of rabies by mouse inoculation. Measurement of humoral immunity to rabies by mouse protection test. Proc Soc Exp Biol Med 32:570–573. https://doi.org/10.3181/00379727-32-7767P
World Health Organization (2013) WHO Expert Consultation on rabies. https://apps.who.int/iris/handle/10665/85346
World Health Organization (2018) WHO Expert Consultation on rabies. https://apps.who.int/iris/handle/10665/272364
Acknowledgments
We want to thank Dr. Jesica Risiglione from the “Instituto de Zoonosis Luis Pasteur” of Buenos Aires city, Argentina, for her disinterested help and for giving us human sera to carry out this investigation. Thanks to Eugenia Dalla Fontana for her help in the language correction of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Research involving human participants and/or animals
Informed consent was obtained from all volunteers according to the ethical guidelines for human subject research. The project was approved by the ethical committee from Facultad de Bioquímica y Ciencias Biológicas of the Universidad Nacional del Litoral, Santa Fe, Argentina, and with the endorsement of the National Network of Rabies Diagnostic Laboratories (Argentina).
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(PDF 1151 kb)
Rights and permissions
About this article
Cite this article
Fontana, D., Rodriguez, M.C., Garay, E. et al. Optimization and validation of a blocking ELISA for quantitation of anti-rabies immunoglobulins in multispecies sera. Appl Microbiol Biotechnol 104, 4127–4139 (2020). https://doi.org/10.1007/s00253-020-10490-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-020-10490-6