Abstract
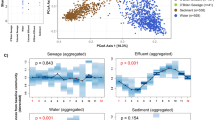
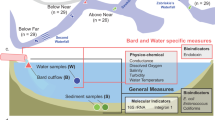
Understanding the microbial quality of recreational waters is critical to effectively managing human health risks. In recent years, the development of new molecular methods has provided scientists with alternatives to the use of culture-based fecal indicator methods for investigating sewage contamination in recreational waters. Before these methods can be formalized into guidelines, however, we must investigate their utility, including strengths and weaknesses in different environmental media. In this study, we investigated the decay of sewage-associated bacterial communities in water and sediment from three recreational areas in Southeast Queensland, Australia. Outdoor mesocosms with water and sediment samples from two marine and one freshwater sites were inoculated with untreated sewage and sampled on days 0, 1, 4, 8, 14, 28, and 50. Amplicon sequencing was performed on the DNA extracted from water and sediment samples, and SourceTracker was used to determine the decay of sewage-associated bacterial communities and how they change following a contamination event. No sewage-associated operational taxonomic units (OTUs) were detected in water and sediment samples after day 4; however, the bacterial communities remained changed from their background measures, prior to sewage amendment. Following untreated sewage inoculation, the mesocosm that had the most diverse starting bacterial community recovered to about 60% of its initial community composition, whereas the least diverse bacterial community only recovered to about 30% of its initial community composition. This suggests that a more diverse bacterial community may play an important role in water quality outcomes after sewage contamination events. Further investigation into potential links between bacterial communities and measures of fecal indicators, pathogens, and microbial source tracking (MST) markers is warranted and may provide insight for recreational water decision-makers.




Similar content being viewed by others
References
Ahmed W, Gyawali P, Sidhu JPS, Toze S (2014) Relative inactivation of faecal indicator bacteria and sewage markers in freshwater and seawater microcosms. J Appl Micrbiol 59:348–354
Ahmed W, Staley C, Sadowsky MJ, Gyawali P, Sidhu JPS, Palmer A (2015) Toolbox approaches using molecular markers and 16S rRNA gene amplicon data sets for identification of fecal pollution in surface water. Appl Environ Microbiol 81:7067–7077
Ahmed W, Hamilton K, Gyawali P, Toze S, Haas CN (2016a) Evidence of avian and possum fecal contamination in rainwater tanks as determined by microbial source tracking approaches. Appl Environ Microbiol 82:4379–4386
Ahmed W, Hughes B, Harwood VJ (2016b) Current status of marker genes of Bacteroides and related taxa for identifying sewage pollution in environmental waters. Water 8:231
Ahmed W, Staley C, Sidhu JPS, Sadowsky M, Toze S (2017) Amplicon-based profiling of bacteria in raw and secondary treated wastewater from treatment plants across Australia. Appl Microbiol Biotechnol 101:1253–1266
Alkan U, Elliot DJ, Evison M (1995) Survival of enteric bacteria in relation to simulated solar radiation and other environmental factors in marine waters. Water Res 29:2071–2080
Anderson MJ, Willis TJ (2003) Canonical analysis of principal coordinates: a useful method of constrained ordination for ecology. Ecology 84:511–525
Anderson KL, Whitlock JE, Harwood VJ (2005) Persistence and differential survival of fecal indicator bacteria in subtropical waters and sediments. Appl Environ Microbiol 71:3041–3048
Arnold BF, Wade TJ, Benjamin-Chung J, Schiff KC, Griffith JF, Dufour AP, Weisberg SB, Colford JM Jr (2016) Acute gastroenteritis and recreational water: highest burden among young US children. Am J Public Health 106:1690–1697
Aronesty E (2013) Comparison of sequencing utility programs. Open Bioinform J 7:1–8
Bae S, Wuertz S (2012) Survival of host-associated Bacteroides cells and their relationship with Enterococcus spp., Campylobacter jejuni, Salmonella enterica serovar Typhimurium, and adenovirus in freshwater microcosms as measured by propidium monoazide-quantitative PCR. Appl Environ Microbiol 78:922–932
Beale DJ, Crosswell J, Karpe AV, Ahmed W, Williams M, Morrison PD, Metcalfe S, Staley C, Sadowsky MJ, Palombo EA, Steven A (2017) A multi-omics based ecological analysis of coastal marine sediments from Gladstone, in Australia’s Far North Queensland, and Heron Island, a nearby fringing platform reef. Sci Total Environ 609:842–853
Bradshaw JK, Snyder BJ, Oladeinde A, Spidle D, Berrang ME, Meinersmann RJ, Oakley B, Sidle RC, Sullivan K, Molina M (2016) Charectrizing relationships among fecal indicator bacteria, microbial source tracking markers, and associated waterborne pathogen occurrence in stream water and sediments in a mixed land use watershed. Water Res 101:498–509
Bray JR, Curtis JT (1957) An ordination of the upland forest communities of southern Wisconsin. Ecol Monogr 27:325–349
Brown CM, Staley C, Wang P, Dalzell B, Chun CL, Sadowsky MJ (2017) A high-throughput DNA-sequencing approach for determining sources of fecal bacteria in a lake superior estuary. Environ Sci Technol 51:8263–8271
Byappanahalli MN, Nevers MB, Korajkic A, Staley ZR, Harwood VJ (2012) Enterococci in the environment. Microbiol Mol Biol Rev 76:685–706
Claesson MJ, Wang QO, O’Sullivan O, Greene-Diniz R, Cole JR, Ross RP, O’Toole PW (2010) Comparison of two next-generation sequencing technologies for resolving highly complex microbiota composition using tandem variable 16S rRNA gene regions. Nucleic Acids Res 38:e200
Clarke KR (1993) Non-parametric multivariate analyses of changes in community structure. Aust J Ecol 18:117–143
Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM (2009) The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res 37:D141–D145
de Abreu CA, Souza DS, Moresco V, Kleemann CR, Garcia LA, Barardi CR (2012) Stability of human enteric viruses in seawater samples from molluscs depuration tanks coupled with ultraviolet irradiation. J Appl Microbiol 113:1554–1563
Dick LK, Stelzer EA, Bertke EE, Fong DL, Stoeckel DM (2010) Relative decay of Bacteroidales microbial source tracking markers and cultivated Escherichia coli in freshwater microcosms. Appl Environ Microbiol 76:3255–3262
Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200
Eichmiller JJ, Borchert AJ, Sadowsky MJ (2014) Decay of genetic markers for fecal bacterial indicators and pathogens in sand from lake superior. Water Res 59:99–111
Fujioka RS, Solo-Gabriele HM, Byappanahalli MN, Kirs M (2015) U.S. recreational water quality criteria: a vision for the future. Int J Environ Res Public Health 12:7752–7776
Garcia-Armisen T, Inceoglu O, Ouattara NK, Anzil A, Verbanck MA, Brion N, Servais P (2014) Seasonal variations and resilience of bacterial communities in a sewage polluted urban river. PLoS One 9:e92579
Gihring TM, Green SJ, Schadt CW (2012) Massively parallel rRNA gene sequencing exacerbates the potential for biased community diversity comparisons due to variable library sizes. Environ Microbiol 14:285–290
Gohl DM, Vangay P, Garbe J, MacLean A, Hauge A, Becker A, Gould TJ, Clayton JB, Johnson TJ, Hunter R, Knights D, Beckman KB (2016) Systematic improvement of amplicon marker gene methods for increased accuracy in microbiome studies. Nat Biotechnol 34:942–949
Green HC, Shanks OC, Sivaganesan M, Haugland RA, Field KG (2011) Differential decay of human faecal Bacteroides in marine and freshwater. Environ Microbiol 13:3235–3249
Halliday E, Gast RJ (2011) Bacteria in beach sands: an emerging challenge in protecting coastal water quality and bather health. Environ Sci Technol 45:370–379
Harwood VJ, Staley C, Badgely BD, Borges K, Korajkic A (2014) Microbial source tracking markers for detection of fecal contamination in environmental waters: relationships between pathogens and human health outcomes. FEMS Microbiol Rev 38:1–40
Haugland RA, Siefring SC, Wymer LJ, Brenner KP, Dufour AP (2005) Comparison of Enterococcus measurements in freshwater at two recreational beaches by quantitative polymerase chain reaction and membrane filter culture analysis. Water Res 39:559–568
Henry R, Schang C, Coutts S, Kolotelo P, Prosser T, Crosbie N, Grant T, Cottam D, O’Brien P, Deletic A, McCarthy D (2016) Into the deep: evaluation of SourceTracker for assessment of faecal contamination of coastal waters. Water Res 93:242–253
Huse SM, Welch DM, Morrison HG, Sogin ML (2010) Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environ Microbiol 12:889–898
Ishii S, Sadowsky MJ (2008) Escherichia coli in the environment: implications for water quality and human health. Microbes Environ 23:101–108
Ishii S, Ksoll WB, Hicks RE, Sadowsky MJ (2006) Presence and growth of naturalized Escherichia coli in temperate soils from Lake Superior waterheds. Appl Environ Microbiol 72:612–621
Knights D, Kuczynski J, Charlson ES, Zaneveld J, Mozer MC, Collman RG, Bushman FD, Knight R, Kelly ST (2011) Bayesian community-wide culture-independent microbial source tracking. Nat Methods 8:761–763
Korajkic A, Wanjugi P, Harwood VJ (2013) Indigenous microbiota and habitat influence Escherichia coli survival more than sunlight in simulated aquatic environments. Appl Environ Microbiol 79:5329–5337
Korajkic A, Parfrey LW, McMinn BR, Baeza YV, VanTeuren W, Knight R, Shanks OC (2015) Changes in bacterial and eukaryotic communities during sewage decomposition in Mississippi River water. Water Res 69:30–39
Kunin V, Engelbrektson A, Ochman H, Hugenholtz P (2010) Wrinkles in the rare biosphere: pyrosequencing errors can lead to artificial inflation of diversity estimates. Environ Microbiol 12:118–123
Liang Z, He Z, Zhou X, Powell CA, Yang Y, Roberts MG, Stoffella PJ (2012) High diversity and differential decay persistence of fecal Bacteroidales population spiked into freshwater microcosm. Water Res 46:247–257
Lu S, Sun Y, Zhao X, Wang L, Ding A, Zhao X (2016) Sequencing insights into microbial communities in the water and sediments of Fenghe River, China. Arch Environ Contam Toxicol 71:122–132
Madoux-Humery AS, Dorner S, Sauve S, Aboulfadl K, Galarneau M, Servais P, Prevost M (2013) Temporal variability of combines sewer overflow contaminants: evaluation of wastewater micropollutants as tracers of fecal contamination. Water Res 47:4370–4282
Mandaric L, Mor JR, Sabater S, Petrovic M (2017) Impact of urban chemical pollution on water quality in small, rural and effluent-dominated Mediterranean streams and rivers. Sci Total Environ 613-614:763–772
Mattioli MC, Sassoubre LM, Russel TL, Boehm AB (2017) Decay of sewage-sourced microbial source tracking markers and fecal indicator bacteria in marine waters. Water Res 108:106–114
Murphu HM, Thomas MK, Schmidt PJ, Medeiros DT, McFadyen S, Pintar KD (2016) Estimating the burden of acute gastrointestinal illness due to Giardia, Cryptosporidium, Campylobacter, E. coli O157 and norovirus associated with private wells and small water systems in Canada. Epidemiol Infect 144:1355–1370
Naidoo S, Olaniran AO (2014) Treated wastewater effluent as a source of microbial pollution of surface water resources. Int J Environ Res Public Health 11:249–270
Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig WG, Peplies J, Glockner FO (2007) SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35:7188–7196
Richards S, Withers PJ, Paterson E, McRoberts CW, Stutter M (2016) Temporal variability in domestic point source discharges and their associated impact on receiving waters. Sci Total Environ 571:1275–1283
Rochelle-Newall E, Nguyen TM, Le TP, Sengtaheuanghpung O, Ribolzi O (2015) A short review of fecal indicator bacteria in tropical aquatic ecosystems: knowledge gaps and future directions. Front Microbiol 6:308
Sassoubre LM, Yamahara KM, Boehm AB (2015) Temporal stability of the microbial community in sewage-polluted seawater exposed to natural sunlight cycles and marine microbiota. Appl Environ Microbiol 81:2107–2116
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541
Schulz CJ, Childers GW (2011) Faecal Bacteroidales diversity and decay in response to variations in temperature and salinity. Appl Environ Microbiol 77:2563–2572
Shuval H (2003) Estimating the global burden of thalassogenic diseases: human infectious diseases caused by wastewater pollution in the environment. J Water Health 1:53–64
Sinton LW, Hall CH, Lynch PA, Davies-Colley RJ (2002) Sunslight inactivation of fecal indicator bacteria and bacteriophages from waste stabilization pond effluent in fresh and saline waters. Appl Environ Microbiol 68:1122–1131
Sogin ML, Morrison HG, Huber JA, Mark Welch D, Huse SM, Neal PR, Arrieta JM, Herndl GJ (2006) Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proc Natl Acad Sci U S A 103:12115–20
Staley C, Gordon KV, Schoen ME, Harwood VJ (2012) Performance of two quantitative PCR methods for microbial source tracking of human sewage and implications for microbial risk assessment in recreational waters. Appl Environ Microbiol 78:7317–7326
Staley ZR, Grabuski J, Sverko E, Edge TA (2016) Comparison of microbial and chemical source tracking markers to identify fecal contamination sources in the Humber River (Toronto, Ontario, Canada) and associated storm water outfalls. Appl Environ Microbiol 82:6357–6366
Staley C, Kaiser T, Gidley ML, Enochs IC, Jones PR, Goodwin KD, Sinigalliano CD, Sadowsky MJ, Chun CL (2017) Differential impacts of land-based sources of pollution on the microbiota of Southeast Florida Coral Reefs. Appl Environ Microbiol 83:e03378–e03316
Staley C, Kaiser T, Lobos A, Ahmed W, Harwood VJ, Brown CM, Sadowsky MJ (2018) Application of SourceTracker for accurate identification of fecal pollution in recreational freshwater: a double-blinded study. Environ Sci Technol 52:4207–4217
US Environmental Protection Agency (2012) Recreational Water Quality Criteria. Health and Ecological Criteria Division, Office of Science and Technology, United States Environmental Protection Agency, Washington, D.C
Walters SP, Yamahara KM, Boehm AB (2009) Persistence of nucleic acid markers of health-relevant organisms in seawater microcosms: implications for their use in assessing risk in recreational waters. Water Res 43:4929–4939
Whitman RL, Nevers MB, Korinek GC, Byappanahalli MN (2004) Solar and temporal effects on Escherichia coli concentration at a Lake Michigan Swimming Beach. Appl Environ Microbiol 70:4276–4285
Winterbourn JB, Clements K, Lowther JA, Malhalm SK, McDonald JE, Jones DL (2016) Use of Mytilus edulis biosentinels to investigate spatial patterns of norovirus and faecal indicator organism contamination around coastal sewage discharges. Water Res 105:241–250
Wolf L, Held I, Eiswirth M, Hotzl H (2004) Impact of leaky sewers on groundwater quality. Clean Soil Air Water 32:361–373
World Health Organization (2003) Guidelines for safe recreational water environments. World Health Organization, Geneva
Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, Heath AC, Warner B, Reeder J, Kuczynski J, Caporaso JG, Lozupone CA, Lauber C, Clemente JC, Knights D, Knight R, Gordon JI (2012) Human gut microbiome viewed across age and geography. Nature 486:222−227
Zhang Q, He X, Yan T (2015) Differential decay of wastewater bacteria and change of microbial communities in beach sand and seawater microcosms. Environ Sci Technol 49:8531–8540
Zhang Q, Eichmiller JJ, Staley C, Sadowsky MJ, Ishii S (2016) Correlations between pathogen concentration and fecal indicator marker genes in beach environments. Sci Total Environ 573:826–830
Acknowledgements
Our thanks to CSIRO Land and Water for funding this strategic project. Sequence processing and analyses were done using the resources of the Minnesota Supercomputing Institute.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical statement
This article does not contain any studies with human participants or animals performed by any of the authors.
Rights and permissions
About this article
Cite this article
Ahmed, W., Staley, C., Kaiser, T. et al. Decay of sewage-associated bacterial communities in fresh and marine environmental waters and sediment. Appl Microbiol Biotechnol 102, 7159–7170 (2018). https://doi.org/10.1007/s00253-018-9112-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-018-9112-4