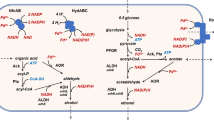
Abstract
Methanol utilization by methylotrophic or non-methylotrophic organisms is the first step toward methanol bioconversion to higher carbon-chain chemicals. Methanol oxidation using NAD-dependent methanol dehydrogenase (Mdh) is of particular interest because it uses NAD+ as the electron carrier. To our knowledge, only a limited number of NAD-dependent Mdhs have been reported. The most studied is the Bacillus methanolicus Mdh, which exhibits low enzyme specificity to methanol and is dependent on an endogenous activator protein (ACT). In this work, we characterized and engineered a group III NAD-dependent alcohol dehydrogenase (Mdh2) from Cupriavidus necator N-1 (previously designated as Ralstonia eutropha). This enzyme is the first NAD-dependent Mdh characterized from a Gram-negative, mesophilic, non-methylotrophic organism with a significant activity towards methanol. Interestingly, unlike previously reported Mdhs, Mdh2 does not require activation by known activators such as B. methanolicus ACT and Escherichia coli Nudix hydrolase NudF, or putative native C. necator activators in the Nudix family under mesophilic conditions. This enzyme exhibited higher or comparable activity and affinity toward methanol relative to the B. methanolicus Mdh with or without ACT in a wide range of temperatures. Furthermore, using directed molecular evolution, we engineered a variant (CT4-1) of Mdh2 that showed a 6-fold higher K cat/K m for methanol and 10-fold lower K cat/K m for n-butanol. Thus, CT4-1 represents an NAD-dependent Mdh with much improved catalytic efficiency and specificity toward methanol compared with the existing NAD-dependent Mdhs with or without ACT activation.







Similar content being viewed by others
References
Arfman N, Watling EM, Clement W, van Oosterwijk RJ, de Vries GE, Harder W, Attwood MM, Dijkhuizen L (1989) Methanol metabolism in thermotolerant methylotrophic Bacillus strains involving a novel catabolic NAD-dependent methanol dehydrogenase as a key enzyme. Arch Microbiol 152(3):280–288. doi:10.1007/BF00409664
Arfman N, Van Beeumen J, De Vries GE, Harder W, Dijkhuizen L (1991) Purification and characterization of an activator protein for methanol dehydrogenase from thermotolerant Bacillus spp. J Biol Chem 266(6):3955–3960
Arfman N, Hektor HJ, Bystrykh LV, Govorukhina NI, Dijkhuizen L, Frank J (1997) Properties of an NAD(H)-containing methanol dehydrogenase and its activator protein from Bacillus methanolicus. Eur J Biochem 244(2):426–433. doi:10.1111/j.1432-1033.1997.00426.x
Bogorad IW, Lin T-S, Liao JC (2013) Synthetic non-oxidative glycolysis enables complete carbon conservation. Nature 502(7374):693–7. doi:10.1038/nature12575
Bogorad IW, Chen C-T, Theisen MK, Wu T-Y, Schlenz AR, Lam AT, Liao JC (2014) Building carbon-carbon bonds using a biocatalytic methanol condensation cycle. Proc Natl Acad Sci U S A 111(45):15928–15933. doi:10.1073/pnas.1413470111
Carpenter EP, Hawkins AR, Frost JW, Brown KA (1998) Structure of dehydroquinate synthase reveals an active site capable of multistep catalysis. Nature 394(6690):299–302. doi:10.1038/28431
De Vries GE, Arfman N, Terpstra P, Dijkhuizen L (1992) Cloning, expression, and sequence analysis of the Bacillus methanolicus C1 methanol dehydrogenase gene. J Bacteriol 174(16):5346–5353
Elleuche S, Antranikian G (2013) Bacterial group III alcohol dehydrogenases—function, evolution and biotechnological applications. OA Alcohol 1(1):1–6. doi:10.13172/2053-0285-1-1-489
Gibson DG, Young L, Chuang R-Y, Venter JC, Hutchison CA III, Smith HO (2009) Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6(5):343–345. doi:10.1038/nmeth.1318
Guex N, Peitsch MC, Schwede T (2009) Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: a historical perspective. Electrophoresis 30(S1):S162–S173. doi:10.1002/elps.200900140
Hagishita T, Yoshida T, Izumi Y, Mitsunaga T (1996) Efficient L-serine production from methanol and glycine by resting cells of Methylobacterium sp. strain MN43. Biosci Biotechnol Biochem 60(10):1604–1607. doi:10.1271/bbb.60.1604
Hektor HJ, Kloosterman H, Dijkhuizen L (2002) Identification of a magnesium-dependent NAD(P)(H)-binding domain in the nicotinoprotein methanol dehydrogenase from Bacillus methanolicus. J Biol Chem 277(49):46966–46973. doi:10.1074/jbc.M207547200
Keltjens JT, Pol A, Reimann J, Op den Camp HJM (2014) PQQ-dependent methanol dehydrogenases: rare-earth elements make a difference. Appl Microbiol Biotechnol 98(14):6163–6183. doi:10.1007/s00253-014-5766-8
Kim YH, Campbell E, Yu J, Minteer SD, Banta S (2013) Complete oxidation of methanol in biobattery devices using a hydrogel created from three modified dehydrogenases. Angew Chem Int Ed 52(5):1437–1440. doi:10.1002/anie.201207423
Kloosterman H, Vrijbloed JW, Dijkhuizen L (2002) Molecular, biochemical, and functional characterization of a Nudix hydrolase protein that stimulates the activity of a nicotinoprotein alcohol dehydrogenase. J Biol Chem 277(38):34785–34792. doi:10.1074/jbc.M205617200
Kotrbova-Kozak A, Kotrba P, Inui M, Sajdok J, Yukawa H (2007) Transcriptionally regulated adhA gene encodes alcohol dehydrogenase required for ethanol and n-propanol utilization in Corynebacterium glutamicum R. Appl Microbiol Biotechnol 76(6):1347–1356. doi:10.1007/s00253-007-1094-6
Krog A, Heggeset TMB, Müller JEN, Kupper CE, Schneider O, Vorholt JA, Ellingsen TE, Brautaset T (2013) Methylotrophic Bacillus methanolicus encodes two chromosomal and one plasmid born NAD+ dependent methanol dehydrogenase paralogs with different catalytic and biochemical properties. PLoS One 8(3):e59188. doi:10.1371/journal.pone.0059188
Lutz R, Bujard H (1997) Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res 25(6):1203–1210. doi:10.1093/nar/25.6.1203
Mani J-C, Pietruszko R, Theorell H (1970) Methanol activity of alcohol dehydrogenases from human liver, horse liver, and yeast. Arch Biochem Biophys 140(1):52–59. doi:10.1016/0003-9861(70)90009-3
Marçal D, Rêgo AT, Carrondo MA, Enguita FJ (2009) 1,3-Propanediol dehydrogenase from Klebsiella pneumoniae: decameric quaternary structure and possible subunit cooperativity. J Bacteriol 191(4):1143–1151. doi:10.1128/JB.01077-08
Montella C, Bellsolell L, Pérez-Luque R, Badía J, Baldoma L, Coll M, Aguilar J (2005) Crystal structure of an iron-dependent group III dehydrogenase that interconverts L-lactaldehyde and L-1,2-propanediol in Escherichia coli. J Bacteriol 187(14):4957–4966. doi:10.1128/JB.187.14.4957
Moon J-H, Lee H-J, Park S-Y, Song J-M, Park M-Y, Park H-M, Sun J, Park J-H, Kim BY, Kim J-S (2011) Structures of iron-dependent alcohol dehydrogenase 2 from Zymomonas mobilis ZM4 with and without NAD+ cofactor. J Mol Biol 407(3):413–424. doi:10.1016/j.jmb.2011.01.045
Motoyama H, Anazawa H, Katsumata R, Araki K, Teshiba S (1993) Amino acid production from methanol by Methylobacillus glycogenes mutants: isolation of L-glutamic acid hyper-producing mutants from M. glycogenes strains, and derivation of L-threonine and L-lysine-producing mutants from them. Biosci Biotechnol Biochem 57(1):82–87. doi:10.1271/bbb.57.82
Motoyama H, Yano H, Ishino S, Anazawa H, Teshiba S (1994) Effects of the amplification of the genes coding for the L-threonine biosynthetic enzymes on the L-threonine production from methanol by a gram-negative obligate methylotroph, Methylobacillus glycogenes. Appl Microbiol Biotechnol 42(1):67–72. doi:10.1007/s002530050218
Motoyama H, Yano H, Terasaki Y, Anazawa H (2001) Overproduction of L-lysine from methanol by Methylobacillus glycogenes derivatives carrying a plasmid with a mutated dapA Gene. Appl Environ Microbiol 67(7):3064–3070. doi:10.1128/AEM.67.7.3064-3070.2001
Müller JEN, Meyer F, Litsanov B, Kiefer P, Potthoff E, Heux S, Quax WJ, Wendisch VF, Brautaset T, Portais J-C, Vorholt JA (2015) Engineering Escherichia coli for methanol conversion. Metab Eng 28:190–201. doi:10.1016/j.ymben.2014.12.008
Nash T (1953) The colorimetric estimation of formaldehyde by means of the Hantzsch reaction. Biochem J 55(3):416–421
Notredame C, Higgins DG, Heringa J (2000) T-Coffee: a novel method for fast and accurate multiple sequence alignment. J Mol Biol 302(1):205–217. doi:10.1006/jmbi.2000.4042
Ochsner AM, Müller JEN, Mora CA, Vorholt JA (2014) In vitro activation of NAD-dependent alcohol dehydrogenases by Nudix hydrolases is more widespread than assumed. FEBS Lett 588(17):2993–2999. doi:10.1016/j.febslet.2014.06.008
Robert X, Gouet P (2014) Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res 42(W1):W320–W324. doi:10.1093/nar/gku316
Ruzheinikov SN, Burke J, Sedelnikova S, Baker PJ, Taylor R, Bullough PA, Muir NM, Gore MG, Rice DW (2001) Glycerol dehydrogenase: structure, specificity, and mechanism of a family III polyol dehydrogenase. Structure 9(9):789–802. doi:10.1016/S0969-2126(01)00645-1
Schrader J, Schilling M, Holtmann D, Sell D, Filho MV, Marx A, Vorholt JA (2009) Methanol-based industrial biotechnology: current status and future perspectives of methylotrophic bacteria. Trends Biotechnol 27(2):107–115. doi:10.1016/j.tibtech.2008.10.009
Sheehan MC, Bailey CJ, Dowds BCA, McConnell DJ (1988) A new alcohol dehydrogenase, reactive towards methanol, from Bacillus stearothermophilus. Biochem J 252(3):661–666
The Uniprot Consortium (2015) UniProt: a hub for protein information. Nucleic Acids Res 43(D):D204–D212. doi:10.1093/nar/gku989
Whitaker WB, Sandoval NR, Bennett RK, Fast AG, Papoutsakis ET (2015) Synthetic methylotrophy: engineering the production of biofuels and chemicals based on the biology of aerobic methanol utilization. Curr Opin Biotechnol 33:165–175. doi:10.1016/j.copbio.2015.01.007
Wierenga RK, Terpstra P, Hol WGJ (1986) Prediction of the occurrence of the ADP-binding beta alpha beta-fold in proteins, using an amino acid sequence fingerprint. J Mol Biol 187(1):101–107. doi:10.1016/0022-2836(86)90409-2
Witthoff S, Mühlroth A, Marienhagen J, Bott M (2013) C1 metabolism in Corynebacterium glutamicum: an endogenous pathway for oxidation of methanol to carbon dioxide. Appl Environ Microbiol 79(22):6974–6983. doi:10.1128/AEM.02705-13
Witthoff S, Schmitz K, Niedenführ S, Nöh K, Noack S, Bott M, Marienhagen J (2015) Metabolic engineering of Corynebacterium glutamicum for methanol metabolism. Appl Environ Microbiol 81(6):2215–2225. doi:10.1128/AEM.03110-14
Zhang J-H, Chung TDY, Oldenburg KR (1999) A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen 4(2):67–73
Acknowledgments
We are grateful to members of Liao laboratory Matthew C. Siracusa, Saro Avedikian, Tuan Trinh, and Jessica Han Pham for experiment assistance.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Compliance with ethical standards
This article does not contain any studies with human participants or animals performed by any of the authors.
Funding
This work is supported by the Reducing Emissions using Methanotrophic Organisms for Transportation Energy (REMOTE) program of the Advanced Research Projects Agency-Energy (Award: DE- AR0000430). This material is based upon research performed in a renovated collaboratory by the National Science Foundation under Grant No. 0963183, which is an award funded under the American Recovery and Reinvestment Act of 2009 (ARRA).
Conflict of interest
All the authors declare that he/she has no conflict of interest.
Additional information
Tung-Yun Wu and Chang-Ting Chen contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 203 kb)
Rights and permissions
About this article
Cite this article
Wu, TY., Chen, CT., Liu, J.TJ. et al. Characterization and evolution of an activator-independent methanol dehydrogenase from Cupriavidus necator N-1. Appl Microbiol Biotechnol 100, 4969–4983 (2016). https://doi.org/10.1007/s00253-016-7320-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-016-7320-3