Abstract
The VP2 protein of bluetongue virus (BTV) is an important structural protein and is the principal antigen responsible for BTV serotype specificity. In this study, we mapped the reactivity of two BTV16-specific monoclonal antibodies (MAbs) and identified two novel serotype-specific linear B cell epitopes on the BTV16 VP2 protein. By screening a series of peptides derived from the BTV16 VP2 protein and expressed as mannose-binding protein fusions, we determined that the linear epitopes recognized by the VP2-specific MAbs 3 G10 and 2B4 were located within the peptides 34EWSGHDVTEIPNRRMF49 and 540KNEDPYVKRTVKPIRA555, respectively. To define the minimal region required for antibody binding within these peptide regions, a series of progressively shorter peptides were synthesized and evaluated for 3 G10 and 2B4 binding. This work defined the motifs 34EWSGHDVTEIPNRRMF49 and 543DPYVKRTVK555 as the minimal linear peptides required for 3 G10 and 2B4 binding, respectively. Alignment of amino acid sequences from a number of BTV16 strains isolated from different regions indicated that these two epitopes are highly conserved among BTV16 strains. Furthermore, these two epitopes are not conserved among other BTV serotypes or prototype members of the genus Orbivirus in the family Reoviridae, as shown by sequence alignments. The MAb reagents and linear epitopes defined here provide the basis for the development of epitope-based serotype-specific differential diagnostic tools and may be useful in the design of epitope-based vaccines.
Similar content being viewed by others
Introduction
Bluetongue virus (BTV) is the etiological agent of bluetongue disease, a noncontagious, insect-transmitted disease of sheep and some species of wild ruminants (Boone et al. 2007). Due to the potential rapid spread of BTV and the threat to the international trade of livestock and livestock products, bluetongue disease was listed as 1 of 16 diseases formerly classified in list A by the Office International Des Epizooties.
BTV is the prototype member of the genus Orbivirus, family Reoviridae. The BTV genome consists of ten double-stranded RNA genome segments that collectively encode seven structural (VP1 to VP7) and five nonstructural (NS1, NS2, NS3, NS3A, and NS4) proteins (Roy 1989, 1992; Ratinier et al. 2011). The inner capsid or core is composed of two major proteins (VP3 and VP7) and three minor proteins (VP1, VP4, and VP6) and contains the viral genome (Roy 1992, 2005). VP2 and VP5 proteins form a continuous layer over the inner capsid in which multimers of VP2 (dimers and/or trimers) are layered upon a VP5 scaffold. The resulting structures protrude 3 nm beyond the main body of the virion particle and are responsible for the adsorption and entry of BTV into mammalian cells, are responsible for hemagglutination, and are the target of neutralizing antibodies and are, therefore, the primary determinants of serotype specificity (Roy 2008; Hassan and Roy 1999; Bonneau et al. 2001; Zhang et al. 2010). There is considerable genetic and antigenic variation within the BTV serogroup, with 26 distinct serotypes and considerable strain variation within each serotype. As a result, there is little or no immune cross-protection among different BTV serotypes and it has been difficult to develop a useful multivalent vaccine. The development of serotype-specific differential diagnostic approaches is currently needed to facilitate the control of BTV.
Virus infection elicits the production of antibodies against viral proteins. Defined B cell epitopes, defined as regions on the surface of the native antigen that are recognized by B cell receptors or specific antibodies, have emerged as promising targets for vaccines and diagnostic reagents (Tian et al. 2012). Specifically, the identification of serotype-specific epitopes on various proteins of the BTV serogroup would have considerable diagnostic value. While the VP2 protein is a major determinant of viral serotype, serotype-specific epitopes on the VP2 protein of BTV16 have not been reported. We developed two serotype-specific monoclonal antibodies (MAbs) that recognized the VP2 protein of BTV16. Here, we define the linear peptide epitopes recognized by these two MAbs using a series of partially overlapping peptides derived from the BTV16 VP2 protein amino acid sequence to map MAb reactivity. These MAbs and their defined linear epitopes may support the development of serotype-specific differential diagnostic tools and marker vaccines which enable differentiation of infected from vaccinated animals.
Materials and methods
Cell lines and virus
Baby hamster kidney-21 cells (BHK-21 cells) were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen, Carlsbad, CA, USA) in a humidified atmosphere with 5 % CO2 at 37 °C. All culture media were supplemented with 10 % heat-inactivated fetal bovine serum (GIBCO, Invitrogen) and antibiotics (0.1 mg ml−1 of streptomycin and 100 IU ml−1 of penicillin). Viruses from each BTV serotype (BTV1–24), Ibaraki virus (IBAV), and Chuzan virus (CV) were acquired from the State Key Laboratory of Veterinary Biotechnology, Harbin Veterinary Research Institute, Chinese Academy of Agricultural Sciences and were propagated in our laboratory using standard techniques. The GenBank accession number of BTV1 (strain SZ97/1) and BTV16 (strain BN96/16) is JN848767 and JN671915, respectively, and the remaining viruses have yet not been sequenced. A BTV16-reactive polyclonal antiserum was produced by infecting the sheep experimentally with BTV16 strain BN96/16.
Expression and characterization of recombinant BTV16 VP2 protein
Recombinant VP2 protein was prepared using the Bac-to-Bac® Baculovirus Expression System according to the manufacturer’s instructions (Invitrogen). Briefly, the full-length BTV16 VP2 coding sequence was amplified using the following primer pair: BTV16 VP2-EcoRI-22-40 (5′-ACGAATTCATGGAGGAGCTAGTTATACC-3′) and BTV16 VP2-SalI-2901-2880 (5′-CCGTCGACTTAAATATTTAGAAGCTTCGTG-3′). Primers incorporated restriction sites (italicized) to facilitate cloning of the amplified VP2 sequence into the pFast-Bac™ vector. The recombinant pFastBac™ vector was transformed into competent DH10Bac™ Escherichia coli, which were subsequently plated on triple antibiotic LB plates (kanamycin, gentamicin, and tetracycline) with halogenated indolyl-β-galactoside (BluoGal; Invitrogen). Bacmid DNA was recovered from transformed colonies and verified via polymerase chain reaction. Recombinant baculoviruses were generated by using the Cellfectin® II Reagent to transfect Sf9 cells with recombinant bacmid. Recombinant protein was purified by Ni-nitrilotriacetic acid affinity chromatography according to the manufacturer’s instructions (Qiagen). The recombinant VP2 protein was evaluated by Western blot (WB) analysis using a BTV16-reactive polyclonal antiserum as a primary antibody source along with a donkey antisheep IgG (whole molecule)–peroxidase antibody (Sigma) as a secondary antibody.
Production and characterization of monoclonal antibodies
BTV16 VP2-reactive MAbs were generated as follows. Briefly, 6-week-old female BALB/c mice were immunized with 100 μg per mouse purified recombinant BTV16 VP2 protein mixed with an equal volume of complete Freund’s adjuvant (Sigma, St. Louis, MO, USA). Two booster injections containing purified BTV16 VP2 in an equal volume of Freund’s incomplete adjuvant were given at 2-week intervals. Three days after the final booster, splenocytes were fused with SP2/0 myeloma cells using polyethylene glycol (PEG 4000; Sigma). The hybridoma cells were plated in 96-well plates and cultured in hypoxanthine–aminopterin–thymidine (HAT) selection medium (DMEM containing 20 % fetal bovine serum, 100 g ml−1 streptomycin, 100 IU ml−1 penicillin, 100 mM hypoxanthine, 16 mM thymidine, and 400 mM aminopterin). After 5 days, the medium was removed and replaced with hypoxanthine–thymidine (HT)-DMEM medium (DMEM containing 20 % fetal bovine serum, 100 g ml−1 streptomycin, 100 IU ml−1 penicillin, 100 mM hypoxanthine, and 16 mM thymidine). After selection in HAT or HT media, hybridoma supernatants were screened for reactivity and specificity by indirect enzyme-linked immunosorbent assay (ELISA), WB, and indirect immunofluorescence assay (IFA) as previously described (Wang et al. 2012a, b). This approach yielded two BTV16 VP2-reactive MAbs, which were named 3 G10 and 2B4.
Polypeptide design and expression
To express a set of 87 partially overlapping peptides derived from the amino acid sequence of the BTV16 VP2 protein (Yang et al. 2011), we synthesized pairs of complementary oligonucleotides encoding each 16-amino-acid-long peptide sequence (Table S1). Oligonucleotides were designed so that the adjacent peptides in the series had five amino acid residues in common (e.g., the first peptide in the series corresponded to amino acids 1–16, while the second peptide in the series corresponded to amino acids 12–27). The oligonucleotide pairs were annealed to one another and cloned into the EcoRI and SalI sites of pMAL™-C4x, which places the encoded VP2-derived peptide in frame with mannose-binding protein (MBP) to produce MBP-fused polypeptides. MBP-fused polypeptides in the series were named MBP-VP2-1 to MBP-VP2-87. The recombinant plasmids were transformed into E. coli BL21 (DE3; Novagen, USA). Transformed cells were grown in LB/Amp medium (1 % tryptone, 0.5 % yeast extract, 1 % NaCl; 0.1 mg/ml ampicillin) to an OD600 of approximately 0.5–0.7. Isopropylthio-β-galactoside was added to a final concentration of 0.5 mM (Pharmacia Biosciences) to induce recombinant protein expression and the cells were agitated for an additional 6 h at 37 °C. Bacteria were pelleted at 9,000 × g for 10 min at 4 °C and lysed. Polyacrylamide gel electrophoresis and WB was carried out to characterize the production of each MBP-fused recombinant polypeptide.
Epitope identification
Two BTV16 VP2-reactive MAbs, 2B4 and 3 G10, were screened against the MBP-fused polypeptide series by indirect ELISA and WB. The ELISA was performed using the expressed MBP-fused polypeptides as coating antigen. The MBP tag without a fused polypeptide and a BTV VP7-specific MAb (clone IB7) were used as controls. The initial screen of the 87 overlapping VP2-derived peptides identified 2 regions of VP2 on which to focus a more refined mapping of the MAb-binding epitopes. To define the minimal linear peptide epitope recognized by MAb 2B4 and 3 G10, we synthesized and screened by ELISA a series of progressively truncated polypeptides based on the results of the initial screen.
Homology analysis
To evaluate the conservation of the two identified linear VP2 epitopes among BTV16 isolates, we aligned sequences corresponding to the region encompassing the 2B4 and 3 G10 peptide epitopes from a number of BTV16 strains isolated from different geographic areas using the DNAMAN software (Lynnon BioSoft Inc., USA). Alignments were also performed between the defined epitopes and the corresponding regions of BTV serotypes 1–26 and prototype members of the genus Orbivirus in the family Reoviridae to evaluate potential epitope conservation.
The reactivity of MAbs with the corresponding epitope regions from different BTV16 strains
Analysis of the conservation of the two identified linear VP2 epitopes among BTV16 isolates using amino acid alignments identified some BTV16 isolates in which the linear epitopes were not 100 % conserved. The corresponding epitope regions of different BTV16 strains that were not identical to the two identified BTV16 VP2 epitopes were synthesized and tested for reactivity with the BTV16 VP2-reactive MAbs 3 G10 and 2B4 by indirect ELISA. The GenBank accession number of BTV16 VP2 used in this paper is no. JN671907.
Results
Expression and characterization of recombinant BTV16 VP2 protein
Recombinant BTV16 VP2 protein was expressed and purified using a baculovirus expression system. WB analysis showed the recombinant protein migrated with the expected molecular weight and was recognized by a BTV16-reactive polyclonal sheep antiserum (Fig. 1, lane 1). Therefore, the recombinant VP2 protein produced in insect cells maintained epitopes that are recognized by antibodies elicited during BTV16 infection of sheep and was a suitable antigen preparation for immunization and hybridoma screening.
Recognition of recombinant VP2 by a BTV16-reactive polyclonal antiserum. Samples collected during production of recombinant VP2 proteins were analyzed by WB for the presence of VP2 protein using a BTV16-reactive polyclonal antiserum. Lane 1 insect cell lysates following infection with recombinant baculovirus encoding the BTV16 VP2 protein, lane 2 insect cell lysates following infection with wild-type baculovirus, lane M PageRuler™ Prestained Protein Ladder
Production and characterization of monoclonal antibodies
We used recombinant BTV16 VP2 protein to immunize mice in order to generate BTV VP2-reactive MAbs. Two clonal hybridoma cell lines that produced BTV16 VP2-reactive MAbs were identified by indirect ELISA and named 3 G10 and 2B4. MAb 3 G10 and 2B4 reacted with recombinant BTV16 VP2 protein expressed in insect cells (Fig. 2a, lanes 2 and 6) and native BTV16 VP2 protein expressed during BTV16 infection of BHK-21 cells (Fig. 2a, lanes 3 and 7) by WB. Next, the reactivity of each MAb against different BTV serotype viruses was evaluated. MAb reactivity was assessed by an IFA against BHK-21 cells infected with BTV serotype viruses, IBAV, CV. MAb 3 G10 and 2B4 reacted only with BTV16-infected cells and did not cross-react with cells infected with IBAV, CV, or any other BTV serotype tested (Fig. 2b, and data not shown).
MAbs 3 G10 and 2B4 recognize BTV16 VP2 protein. a The reactivity of MAb 3 G10 (a) and 2B4 (b) was evaluated by WB against insect cell lysates following infection with recombinant baculovirus encoding the BTV16 VP2 protein (lanes 1 and 5), purified recombinant BTV16 VP2 protein (lanes 2 and 6), BTV16-infected BHK-21 cell lysates (lanes 3 and 7), and uninfected BHK-21 cell lysates (lanes 4 and 8). Lane M PageRuler™ Prestained Protein Ladder. b BTV16-infected BHK-21 cells were evaluated by IFA for reactivity with MAb 3 G10 (a), 2B4 (b) (bar, 10 μm), a BTV16-reactive polyclonal antiserum (c), and a negative control serum (d)
Identification of BTV16 VP2 epitopes recognized by MAbs 3 G10 and 2B4
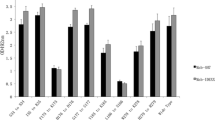
We next evaluated MAb 3 G10 and 2B4 reactivity against a series of 87 partially overlapping peptides derived from the amino acid sequence of the BTV16 VP2 protein in order to map antibody-binding epitopes. Each MBP-fused peptide in the VP2 peptide series was successfully expressed in E. coli and migrated with the expected molecular weight (data not shown). MAb 3 G10 recognized one peptide in the series (MBP-VP2-4), corresponding to the sequence 34EWSGHDVTEIPNRRMF49 of the BTV16 VP2 protein (Fig. 3a). Similarly, MAb 2B4 reacted with one peptide in the series (MBP-VP2-50), corresponding to the sequence 540KNEDPYVKRTVKPIRA555 of the BTV16 VP2 protein (Fig. 3b). Consistent with these results, MAb 3 G10 recognized the MBP-VP2-4 peptide (Fig. 4, lane 1) and MAb 2B4 recognized the MBP-VP2-50 peptide (Fig. 4, lane 3) by WB, but neither antibody reacted with the MBP tag alone (Fig. 4, lanes 2 and 4).
Identification of the BTV16 VP2 linear peptide epitopes recognized by MAb 3 G10 and 2B4. A set of 87 partially overlapping peptides derived from the amino acid sequence of the BTV16 VP2 protein was screened by indirect ELISA to identify linear peptide epitopes recognized by MAb 3 G10 and 2B4. a Screening of MAb 3 G10 against the VP2 peptide series. b Screening of MAb 2B4 against the peptide series. An anti-BTV VP7 MAb was used as a negative control antibody, and the MBP tag with no polypeptide fusion was used as a negative control protein. Error bars reflect the results of three experimental repeats
MAbs 3 G10 and 2B4 recognize their identified peptide epitope by WB. Reactivity of MAb 3 G10 (a) was assessed against the recombinant MBP-VP2-4 polypeptide (34EWSGHDVTEIPNRRMF49, lane 1) and the MBP tag alone (lane 2). Reactivity of MAb 2B4 (b) was assessed against the recombinant MBP-VP2-50 polypeptide (540KNEDPYVKRTVKPIRA555, lane 3) and the MBP tag alone (lane 4). Lane M PageRuler™ Prestained Protein Ladder
We next sought to identify the minimal linear peptide epitopes in the BTV16 VP2 protein recognized by MAb 3 G10 and 2B4. A panel of truncated polypeptides were synthesized in which amino acids were progressively deleted from the amino and/or the carboxy terminus of the 16-amino-acid peptides recognized by 3 G10 and 2B4 in Fig. 3 (34EWSGHDVTEIPNRRMF49 and 540KNEDPYVKRTVKPIRA555, respectively). The deletion of a single residue from either the carboxy or amino terminus of the VP2-4 peptide recognized by 3 G10 resulted in reduced antibody binding, and the antibody binding was completely abrogated when a single residue was deleted from either the carboxy or amino terminus of the VP2-4 peptide (Fig. 5a). Therefore, 34EWSGHDVTEIPNRRMF49 is the minimal linear epitope required for optimal 3 G10 binding (Fig. 5a). In contrast, a number of amino acid residues could be deleted from both the carboxy and amino termini of the VP2-50 peptide without any detectable reduction in 2B4 antibody binding (Fig. 5b). Further removal of amino acids from the peptide sequence 543DPYVKRTVK551 resulted in a complete abrogation of 2B4 antibody binding, defining 543DPYVKRTVK551 as the minimal linear epitope recognized by 2B4 (Fig. 5b).
Progressively truncated peptides define the minimal linear epitope recognized by MAb 3 G10 and 2B4. MAbs 3 G10 (a) and 2B4 (b) were screened against a series of truncated peptides that had amino acid residues progressively deleted from the amino and/or carboxy termini to determine the minimal linear peptide sequence required for MAb binding. The name and sequence of the peptides used are included below the figure. A VP7-specific MAb was used as a negative control. Error bars reflect the results of three experimental repeats
Homology analysis
We then evaluated whether the two BTV16 VP2 minimal linear epitopes were conserved among BTV16 strains isolated from different regions. The amino acid sequences corresponding to the region encompassing these two BTV16 VP2 minimal linear epitopes from a number of BTV16 strains were identified and aligned (Tables 1 and 2). Both linear epitopes were highly conserved among the different BTV16 strains. When regions aligning with the MAb 3 G10 epitope 34EWSGHDVTEIPNRRMF49 were evaluated among the different BTV16 strains, we found two strains containing an amino acid substitution (Table 1). The Japanese BTV16 strain (accession number AB686213) had a lysine at amino acid position 46 in place of an arginine, and the Australian BTV16 strain (accession number JQ086232) had a serine at amino acid position 45 in place of the asparagine. The 3 G10 epitope 34EWSGHDVTEIPNRRMF49 was entirely conserved within the Mediterranean (accession number DQ191259) and European (accession number AM773701) BTV16 strains (Table 1). Similarly, the MAb 2B4 epitope 543DPYVKRTVK551 was well-conserved among the BTV16 strains evaluated. The Australian BTV16 strain harbored an isoleucine in place of a valine at amino acid position 550, and the Japanese BTV16 strain had an arginine in place of a lysine residue at amino acid position 551. The minimal linear epitope 543DPYVKRTVK551 was totally conserved within the Mediterranean and European BTV16 sequences (Table 2).
We next tested whether the MAbs 3 G10 and 2B4 recognized peptides containing the single amino acid substitutions in the corresponding epitope regions for some BTV16 isolates as described previously. Peptides with the single amino acid substitutions were synthesized and tested for reactivity with MAbs 3 G10 and 2B4 by indirect ELISA. The peptides EWSGHDVTEIPNKRMF and EWSGHDVTEIPSRRMF, corresponding to the epitope regions of the Japanese and Australian BTV16 isolates, showed strong reactivity with MAb 3 G10 despite the amino acid substitutions (Fig. 6a). The peptide DPYVKRTVR, corresponding to the epitope region of the Japanese BTV16 isolate, was still recognized by MAb 2B4. In contrast, 2B4 recognition the peptide DPYVKRTIK, corresponding to the epitope region of the Australian BTV16 isolate, was strongly reduced (Fig. 6b).
Reactivity of MAb 3 G10 and 2B4 with epitope regions of different BTV16 isolates which contain an amino acid substitution. a Peptides corresponding to the minimal MAb 3 G10 epitope region of a Japanese and Australian BTV16 isolate were synthesized and evaluated for reactivity with MAb 3 G10. b Peptides corresponding to the minimal MAb 2B4 epitope region of a Japanese and Australian BTV16 isolate were synthesized and evaluated for reactivity with MAb 2B4. The name and sequence of the peptides used are included below the figure. A VP7-specific MAb was used as a negative control. Error bars reflect the results of three experimental repeats
Amino acid alignments were also performed to evaluate the conservation of the two identified epitopes among different BTV serotypes and prototype members of the genus Orbivirus in the family Reoviridae. The MAb 3 G10 epitope was very poorly conserved among other BTV serotype viruses (Fig. 7a) and prototype members of the genus Orbivirus in the family Reoviridae (Fig. 7b). Similarly, the MAb 2B4 epitope was also poorly conserved among other BTV serotype viruses (Fig. 7c) and prototype members of the genus Orbivirus in the family Reoviridae (Fig. 7d). Collectively, these data show that the epitopes recognized by MAb 3 G10 and 2B4 are well-conserved among BTV16 isolates, but are not conserved among other BTV serotypes or related orbiviruses.
Amino acid alignments of the epitope regions from BTV serotype viruses and prototype orbiviruses. The sequences corresponding to the region encompassing the two BTV16 VP2 minimal linear epitopes were identified and aligned for a panel of BTV serotype viruses (including BTV1–26) and some prototype members of the genus Orbivirus in the family Reoviridae. a, b Sequence alignment of the region encompassing the MAb 3 G10 epitope 34EWSGHDVTEIPNRRMF49 for BTV1–26 (a) and prototype members of the genus Orbivirus in the family Reoviridae (b). c, d Sequence alignment of the region encompassing the MAb 2B4 epitope 543DPYVKRTVK555 for BTV1–26 (c) and prototype members of the genus Orbivirus in the family Reoviridae (d). The defined epitopes and their corresponding region in BTV1–26 and selected orbiviruses are boxed
Discussion
In the last decade, BTV has spread extensively to new areas and raised concerns regarding animal health, the farming industry, and international trade (Purse et al. 2005; Wilson and Mellor. 2009). There are currently 26 distinct BTV serotypes. Each BTV serotype induces humoral immune responses that are generally not cross-reactive, thereby having an impact on BTV vaccination strategies. The development of serotype-specific differential diagnostic techniques is needed to facilitate BTV control measures. To address this need, we focused on the VP2 protein. VP2 is the principal antigen responsible for BTV serotype classification and is the primary target of the neutralizing antibodies that confer protection against virulent virus challenge. A detailed analysis of the antibody epitopes within the VP2 protein would promote our understanding of BTV-specific immunity and the development of epitope-based marker vaccines and diagnostic tools for BTV (Peng et al. 2008). In this study, we developed two BTV16-reactive MAbs that recognized BTV16 but did not react with other BTV serotypes, IBAV or CV, and used them to map linear epitopes on the BTV16 VP2 protein.
The MAbs 3 G10 and 2B4 recognized recombinant BTV16 VP2 protein expressed in insect cells using the Bac-to-Bac® Baculovirus Expression System and native BTV16 VP2 protein expressed as a result of BTV16 infection of BHK-21 cells by WB. That antibody recognition of VP2 was maintained in WB suggested that both MAbs recognized linear peptide epitopes within the VP2 protein. We identified the region of VP2 recognized by each MAb by screening a set of 87 partially overlapping peptides derived from the amino acid sequence of the BTV16 VP2 protein, expressed as MBP-fused polypeptides (Sun et al. 2012). This screen identified a region of BTV16 VP2 recognized by MAb 3 G10 and a different region recognized by MAb 2B4.
Having identified a linear region of VP2 recognized by each MAb, we synthesized progressively truncated peptides to define the minimal linear epitopes recognized by 3 G10 and 2B4. Removal of amino acid residue 34E at the amino terminus of the polypeptide 34EWSGHDVTEIPNRRMF49 showed reduced binding reactivity with MAb 3 G10 (OD492 value dropping from 3.499 to 1.254). Similarly, deletion of the 49 F amino acid also exhibited reduced binding reactivity with MAb 3 G10 (OD492 value dropping from 3.499 to 0.974). When both 34E and 49 F were removed, MAb 3 G10 binding was completely abolished, indicating that 34EWSGHDVTEIPNRRMF49 is the minimal linear epitope required for optimal 3 G10 binding and 34E and 49 F are important components of the peptide epitope (Fig. 5a).
The initial peptide screen identified the polypeptide 540KNEDPYVKRTVKPIRA555 as the linear region recognized by the MAb 2B4. A number of amino acids could be deleted from both the amino and carboxy termini of this polypeptide without significant reductions in 2B4 binding (Fig. 5b). All synthesized peptides containing the core 543DPYVKRTVK551 were recognized well by 2B4. Further truncation of the amino acid residue 543D resulted in a complete loss of 2B4 binding, illustrating the importance of the amino acid residue 543D in epitope maintenance (Fig. 5b). This work defined 543DPYVKRTVK551 as the minimal linear peptide recognized by 2B4.
Amino acid sequence alignments of a number of BTV16 strains isolated from different regions indicated that these two linear epitopes are highly conserved among BTV16 strains. The corresponding epitope regions of BTV16 strains containing a single amino acid substitution relative to the reference epitope sequence we identified were synthesized and tested for reactivity with the MAbs by indirect ELISA. Despite the amino acid substitution found within some BTV16 isolates, both BTV16 serotype-specific MAbs recognized the corresponding epitope regions of different BTV16 strains isolated from different regions (Fig. 6). These two BTV16 VP2-specific MAbs and their corresponding binding epitopes can provide a foundation for the establishment of new serotype-specific differential diagnostic approaches.
Mapping epitopes using MAbs has become a powerful tool to study protein structure and has been used to diagnose diseases and design marker vaccines (Sukupolvi-Petty et al. 2010; Kouzmitcheva et al. 2001; Langeveld et al. 1994). In this study, we also used DNASTAR software (http://www.dnastar.com/) to analyze features of the BTV16 VP2 protein (GenBank accession number JN671907). This analysis suggested that the two linear epitopes identified in this study and their peripheral regions have a high degree of hydrophilicity, a high antigenic index, and a positive surface probability (data not shown), suggesting that these epitopes stand a good chance of exposing on the surface of the VP2 tertiary structure. This analysis is consistent with our IFA results which showed that both MAb 3 G10 and 2B4 specifically reacted with BTV16-infected cells (Fig. 2b). These results may provide some insight into the three-dimensional structural properties of the BTV VP2 protein.
In conclusion, we developed two BTV16 VP2-specific MAbs and defined the linear peptide epitopes within the VP2 protein recognized by each antibody. The two BTV16 VP2-specific MAbs and their defined linear peptide epitopes may be used in the establishment of serotype-specific differential diagnostic tools and vaccine development.
References
Bonneau KR, Mullens BA, MacLachlan NJ (2001) Occurrence of genetic drift and founder effect during quasispecies evolution of the VP2 and NS3/NS3A genes of bluetongue virus upon passage between sheep, cattle, and Culicoides sonorensis. J Virol 75:8298–8305
Boone JD, Balasuriya UB, Karaca K, Audonnet JC, Yao J, He L, Nordgren R, Monaco F, Savini G, Gardner IA, Maclachlan NJ (2007) Recombinant canarypox virus vaccine co-expressing genes encoding the VP2 and VP5 outer capsid proteins of bluetongue virus induces high level protection in sheep. Vaccine 25:672–678
Hassan SS, Roy P (1999) Expression and functional characterization of bluetongue virus VP2 protein: role in cell entry. J Virol 73:9832–9842
Kouzmitcheva GA, Petrenko VA, Smith GP (2001) Identifying diagnostic peptides for lyme disease through epitope discovery. Clin Diagn Lab Immun 8:150–160
Langeveld JP, Casal JI, Osterhaus AD, Cortes E, de Swart R, Vela C, Dalsgaard K, Puijk WC, Schaaper WM, Meloen RH (1994) First peptide vaccine providing protection against viral infection in the target animal: studies of canine parvovirus in dogs. J Virol 68:4506–4513
Peng WP, Hou Q, Xia ZH, Chen D, Li N, Sun Y, Qiu HJ (2008) Identification of a conserved linear B-cell epitope at the N-terminus of the E2 glycoprotein of classical swine fever virus by phage-displayed random peptide library. Virus Res 135:267–272
Purse BV, Mellor PS, Rogers DJ, Samuel AR, Mertens PP, Baylis M (2005) Climate change and the recent emergence of bluetongue in Europe. Nat Rev Microbiol 3:171–181
Ratinier M, Caporale M, Golder M, Franzoni G, Allan K, Nunes SF, Armezzani A, Bayoumy A, Rixon F, Shaw A, Palmarini M (2011) Identification and characterization of a novel non-structural protein of bluetongue virus. PLoS Pathog 7:e1002477
Roy P (1989) Bluetongue virus genetics and genome structure. Virus Res 13:179–206
Roy P (1992) Bluetongue virus proteins. J Gen Virol 73:3051–3064
Roy P (2005) Bluetongue virus proteins and particles and their role in virus entry, assembly, and release. Adv Virus Res 64:69–123
Roy P (2008) Bluetongue virus: dissection of the polymerase complex. J Gen Virol 89:1789–1804
Sukupolvi-Petty S, Austin SK, Engle M, Brien JD, Dowd KA, Williams KL, Johnson S, Rico-Hesse R, Harris E, Pierson TC, Fremont DH, Diamond MS (2010) Structure and function analysis of therapeutic monoclonal antibodies against dengue virus type 2. J Virol 84:9227–9239
Sun EC, Zhao J, Liu NH, Yang T, Ma JN, Geng HW, Wang LF, Qin YL, Bu ZG, Yang YH, Lunt RA, Wu DL (2012) Comprehensive mapping of West Nile virus (WNV)- and Japanese encephalitis virus serocomplex-specific linear B-cell epitopes from WNV non-structural protein 1. J Gen Virol 93:50–60
Tian Y, Chen W, Yang Y, Xu X, Zhang J, Wang J, Xiao L, Chen Z (2012) Identification of B cell epitopes of dengue virus 2 NS3 protein by monoclonal antibody. Appl Microbiol Biotechnol. doi:10.1007/s00253-012-4419-z
Wang W-S, Sun E-C, Liu N-H, Yang T, Xu Q-Y, Qin Y-L, Zhao J, Feng Y-F, Li J-P, Wei P, Zhang C-Y, Wu D-L (2012a) Monoclonal antibodies against VP7 of bluetongue virus. Hybridoma (Larchmt) 31:469–472
Wang W-S, Sun E-C, Liu N-H, Yang T, Xu Q-Y, Qin Y-L, Zhao J, Feng Y-F, Li J-P, Wei P, Zhang C-Y, Wu D-L (2012b) Identification of three novel linear B-cell epitopes on the VP5 protein of BTV16. Vet Microbiol. doi:10.1016/j.vetmic.2012.11.042
Wilson AJ, Mellor PS (2009) Bluetongue in Europe: past, present and future. Philosophical transactions of the Royal Society of London. Series B, Biological sciences 364:2669–2681
Yang T, Liu N, Xu Q, Sun E, Qin Y, Zhao J, Wu D (2011) Complete genomic sequence of bluetongue virus serotype 16 from China. J Virol 85:13472
Zhang X, Boyce M, Bhattacharya B, Schein S, Roy P, Zhou ZH (2010) Bluetongue virus coat protein VP2 contains sialic acid-binding domains, and VP5 resembles enveloped virus fusion proteins. P Natl Acad Sci USA 107:6292–6297
Acknowledgments
This study was supported by the Special Fund for Agro-scientific Research in the Public Interest (20120356), Fundamental Research Funds for Central Public Welfare Research Institutes (ZGKJ201105), and Fundamental Research Funds for Central Public Welfare Research Institutes (0302012016).
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Wen-Shi Wang and En-Cheng Sun contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 139 kb)
Rights and permissions
About this article
Cite this article
Wang, WS., Sun, EC., Xu, QY. et al. Identification of two novel BTV16-specific B cell epitopes using monoclonal antibodies against the VP2 protein. Appl Microbiol Biotechnol 97, 5933–5942 (2013). https://doi.org/10.1007/s00253-013-4779-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-013-4779-z