Abstract
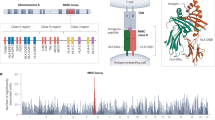
The HLA region of chromosome 6 contains the most polymorphic genes in humans. Spanning ~5 Mbp the densely packed region encompasses approximately 175 expressed genes including the highly polymorphic HLA class I and II loci. Most of the other genes and functional elements are also polymorphic, and many of them are directly implicated in immune function or immune-related disease. For these reasons, this complex genomic region is subject to intense scrutiny by researchers with the common goal of aiding further understanding and diagnoses of multiple immune-related diseases and syndromes. To aid assay development and characterization of the classical loci, a panel of cell lines partially or fully homozygous for HLA class I and II was assembled over time by the International Histocompatibility Working Group (IHWG). Containing a minimum of 88 unique HLA haplotypes, we show that this panel represents a significant proportion of European HLA allelic and haplotype diversity (60–95 %). Using a high-density whole genome array that includes 13,331 HLA region SNPs, we analyzed 99 IHWG cells to map the coordinates of the homozygous tracts at a fine scale. The mean homozygous tract length within chromosome 6 from these individuals is 21 Mbp. Within HLA, the mean haplotype length is 4.3 Mbp, and 65 % of the cell lines were shown to be homozygous throughout the entire region. In addition, four cell lines are homozygous throughout the complex KIR region of chromosome 19 (~250 kbp). The data we describe will provide a valuable resource for characterizing haplotypes, designing and refining imputation algorithms and developing assay controls.






Similar content being viewed by others
References
Coop G, Wen X, Ober C, Pritchard JK, Przeworski M (2008) High-resolution mapping of crossovers reveals extensive variation in fine-scale recombination patterns among humans. Science 319(5868):1395–1398
Coquillard G, Lau M, Kletzel M, Rodriguez-Marino SG (2004) Identification of two pseudogenes with sequence homology to human and gorilla MHC class IA genes: ancestral haplotype in the Filipino population. Hum Immunol 65(6):665–673
Cullen M, Perfetto SP, Klitz W, Nelson G, Carrington M (2002) High-resolution patterns of meiotic recombination across the human major histocompatibility complex. Am J Hum Genet 71(4):759–776
de Bakker PI, Raychaudhuri S (2012) Interrogating the major histocompatibility complex with high-throughput genomics. Hum Mol Genet 21(R1):R29–R36
Dorak MT, Shao W, Machulla HK, Lobashevsky ES, Tang J, Park MH, Kaslow RA (2006) Conserved extended haplotypes of the major histocompatibility complex: further characterization. Genes Immun 7(6):450–467
Gleimer M, Wahl AR, Hickman HD, Abi-Rached L, Norman PJ, Guethlein LA, Hammond JA, Draghi M, Adams EJ, Juo S, Jalili R, Gharizadeh B, Ronaghi M, Garcia KC, Hildebrand WH, Parham P (2011) Although divergent in residues of the peptide binding site, conserved chimpanzee Patr-AL and polymorphic human HLA-A*02 have overlapping peptide-binding repertoires. J Immunol 186(3):1575–1588
Gonzalez-Galarza FF, Takeshita LY, Santos EJ, Kempson F, Maia MH, Silva AL, Ghattaoraya GS, Alfirevic A, Jones AR, Middleton D (2015) Allele frequency net 2015 update: new features for HLA epitopes, KIR and disease and HLA adverse drug reaction associations. Nucleic Acids Res 43(Database issue):D784–D788
Horton R, Wilming L, Rand V, Lovering RC, Bruford EA, Khodiyar VK, Lush MJ, Povey S, Talbot CC Jr, Wright MW, Wain HM, Trowsdale J, Ziegler A, Beck S (2004) Gene map of the extended human MHC. Nat Rev Genet 5(12):889–899
Hosomichi K, Jinam TA, Mitsunaga S, Nakaoka H, Inoue I (2013) Phase-defined complete sequencing of the HLA genes by next-generation sequencing. BMC Genomics 14:355
Illing PT, Vivian JP, Purcell AW, Rossjohn J, McCluskey J (2013) Human leukocyte antigen-associated drug hypersensitivity. Curr Opin Immunol 25(1):81–89
Jeffreys AJ, Kauppi L, Neumann R (2001) Intensely punctate meiotic recombination in the class II region of the major histocompatibility complex. Nat Genet 29(2):217–222
Karolchik D, Barber GP, Casper J, Clawson H, Cline MS, Diekhans M, Dreszer TR, Fujita PA, Guruvadoo L, Haeussler M, Harte RA, Heitner S, Hinrichs AS, Learned K, Lee BT, Li CH, Raney BJ, Rhead B, Rosenbloom KR, Sloan CA, Speir ML, Zweig AS, Haussler D, Kuhn RM, Kent WJ (2014) The UCSC Genome Browser database: 2014 update. Nucleic Acids Res 42(Database issue):D764–D770
Larsen CE, Alford DR, Trautwein MR, Jalloh YK, Tarnacki JL, Kunnenkeri SK, Fici DA, Yunis EJ, Awdeh ZL, Alper CA (2014) Dominant sequences of human major histocompatibility complex conserved extended haplotypes from HLA-DQA2 to DAXX. PLoS Genet 10(10), e1004637
Marsh SGE, Packer R, Heyes JM, Bolton B, Fauchet R, Charron D, Bodmer JG (1996) The International Histocompatibility Workshop cell panel. Genetic diversity of HLA. Functional and medical implications. D. Charron. Paris, EDK. 1: 26–28
Mickelson E, Hurley C, Ng J, Tilanus M, Carrington M, Marsh SGE, Rozemuller E, Pei J, Rosielle J, Voorter C, Sayer D, Damodaran A, Nisperos C, H. JA (2006) 13th IHWS Shared Resources Joint Report. IHWG Cell and Gene Bank and reference cell panels. Immunobiology of the Human MHC: Proceedings of the 13th International Histocompatibilty Workshop and Conference. H. JA. Seattle, IHWG Press. 1: 523–553
Moreno-Estrada A, Gravel S, Zakharia F, McCauley JL, Byrnes JK, Gignoux CR, Ortiz-Tello PA, Martinez RJ, Hedges DJ, Morris RW, Eng C, Sandoval K, Acevedo-Acevedo S, Norman PJ, Layrisse Z, Parham P, Martinez-Cruzado JC, Burchard EG, Cuccaro ML, Martin ER, Bustamante CD (2013) Reconstructing the population genetic history of the Caribbean. PLoS Genet 9(11), e1003925
Myers S, Bottolo L, Freeman C, McVean G, Donnelly P (2005) A fine-scale map of recombination rates and hotspots across the human genome. Science 310(5746):321–324
Parham P, Moffett A (2013) Variable NK cell receptors and their MHC class I ligands in immunity, reproduction and human evolution. Nat Rev Immunol 13(2):133–144
Petersdorf EW (2013) Genetics of graft-versus-host disease: the major histocompatibility complex. Blood Rev 27(1):1–12
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81(3):559–575
Robinson J, Halliwell JA, Hayhurst JD, Flicek P, Parham P, Marsh SG (2015) The IPD and IMGT/HLA database: allele variant databases. Nucleic Acids Res 43(Database issue):D423–D431
Sayers EW (2015) Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 43(Database issue):D6–D17
Steemers FJ, Chang W, Lee G, Barker DL, Shen R, Gunderson KL (2006) Whole-genome genotyping with the single-base extension assay. Nat Methods 3(1):31–33
Stenzel A, Lu T, Koch WA, Hampe J, Guenther SM, De La Vega FM, Krawczak M, Schreiber S (2004) Patterns of linkage disequilibrium in the MHC region on human chromosome 6p. Hum Genet 114(4):377–385
Stewart CA, Horton R, Allcock RJ, Ashurst JL, Atrazhev AM, Coggill P, Dunham I, Forbes S, Halls K, Howson JM, Humphray SJ, Hunt S, Mungall AJ, Osoegawa K, Palmer S, Roberts AN, Rogers J, Sims S, Wang Y, Wilming LG, Elliott JF, de Jong PJ, Sawcer S, Todd JA, Trowsdale J, Beck S (2004) Complete MHC haplotype sequencing for common disease gene mapping. Genome Res 14(6):1176–1187
Tennessen JA, Bigham AW, O'Connor TD, Fu W, Kenny EE, Gravel S, McGee S, Do R, Liu X, Jun G, Kang HM, Jordan D, Leal SM, Gabriel S, Rieder MJ, Abecasis G, Altshuler D, Nickerson DA, Boerwinkle E, Sunyaev S, Bustamante CD, Bamshad MJ, Akey JM (2012) Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science 337(6090):64–69
Terasaki PI (1969) Selection of organ donors. N Engl J Med 280(23):1304
Trowsdale J, Knight JC (2013) Major histocompatibility complex genomics and human disease. Annu Rev Genomics Hum Genet 14:301–323
Watanabe Y, Tokunaga K, Geraghty DE, Tadokoro K, Juji T (1997) Large-scale comparative mapping of the MHC class I region of predominant haplotypes in Japanese. Immunogenetics 46(2):135–141
Williams F, Curran MD, Middleton D (1999) Characterisation of a novel HLA-A pseudogene, HLA-BEL, with significant sequence identity with a gorilla MHC class I gene. Tissue Antigens 54(4):360–369
Yang SY, Milford E, Hammerling U, Dupont B (1987) Description of the reference panel of B-lymphoblastoid cell lines for factors of the HLA system: the B-cell line panel designed for the Tenth International Histocompatibility Wprkshop. Immunobiology of HLA. Histocompatibility Testing. B. Dupont. New York, Springer-Verlag. 1: 11–19
Acknowledgments
We extend thanks to the members of the John Hansen laboratory at the Fred Hutchinson Cancer Research Center for supplying some of the (IHWG) cell line/DNA samples. This study was supported by US National Institutes of Health grant UO1 AI090905.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Figure 1
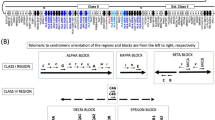
The HLA homozygous cell line panel. a. Columns from left to right: the cell line common names (bold indicates eight cells targeted previously by the MHC sequencing consortium (Horton et al. 2004), IHWG ID numbers, ethnicity, consanguineous (yes/no/unknown) status, HLA-Y presence/absence (Y indicates presence) and classical HLA locus genotype (N/A indicates unknown genotype, red indicates heterozygous at that locus). Data was obtained from the IHWG and IPD websites. Ninety-nine of the cells have IHWG designations and we included one homozygous (WAR) and one heterozygous (EN1NOT) cell line that were not part of the IHWG. (XLSX 26 kb)
Supplementary Figure 2
HLA region homozygous tract coordinates in the IHWC panel. The homozygous tract coordinates were estimated using PLINK (Purcell et al. 2007) and refined manually to show the interval between the last observed heterozygous SNP and the first homozygous SNP in each direction. At the left (Red text) indicates the donor is known to be consanguineous, (Blue text) indicates status unknown. The length of the longest homozygous tract in each cell is shown and (Green text) indicates this extends through the entire HLA region. Over 60% of the cell lines are homozygous through the classical 5.0Mbp HLA region. Shown at the right, four of the cells also have heterozygous tracts that interrupt their homozygous segment. The genome-wide data set is available at ImmPort under SDY295: EXP13576 (https://immport.niaid.nih.gov/). (PDF 401 kb)
Rights and permissions
About this article
Cite this article
Norman, P.J., Norberg, S.J., Nemat-Gorgani, N. et al. Very long haplotype tracts characterized at high resolution from HLA homozygous cell lines. Immunogenetics 67, 479–485 (2015). https://doi.org/10.1007/s00251-015-0857-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00251-015-0857-y