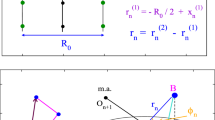
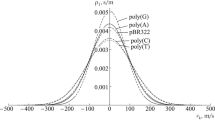
Abstract
Nucleic acids’ physical properties have been investigated by theoretical methods based both on fully atomistic representations and on coarse-grained models, e.g., the worm-like-chain, taken from polymer physics. In this review article, I discuss an intermediate (mesoscopic) approach and show how to build a three-dimensional Hamiltonian model which accounts for the main interactions responsible for the stability of the helical molecules. While the 3D mesoscopic model yields a sufficiently detailed description of the helix at the level of the base pair, it also allows one to predict the thermodynamical and structural properties of molecules in solution. Relying on the idea that the base pair fluctuations can be conceived as trajectories, I have built over the past years a computational method based on the time-dependent path integral formalism to derive the partition function. While the main features of the method are presented, I focus here in particular on a newly developed statistical method to set the maximum amplitude of the base pair fluctuations, a key parameter of the theory. Some applications to the calculation of DNA flexibility properties are discussed together with the available experimental data.









Similar content being viewed by others
Notes
The coefficients \(a_m\) should not be confused with the inverse length \(\bar{a}_n\) of the site-dependent Morse potential.
In principle, the Fourier coefficients in Eq. (11) are integrated on an even domain. However, too negative \(a_m\)’s are discarded due to the physical condition associated to the hard core of the one-particle potential. The latter is tuned by the parameter which regulates the range of the Morse potential (see Section 2). The asymmetry in the choice of \(a_m\)’s included in the computation explains why \(P_j(R_0,\, 0)\) may get slightly larger than 1/2. Hence, the approximation sign is used in the text.
References
Apostolaki A, Kalosakas G (2011) Targets of DNA-binding proteins in bacterial promoter regions present enhanced probabilities for spontaneous thermal openings. Phys Biol 8:026006
Barbi M, Cocco S, Peyrard M (1999) Helicoidal model for DNA opening. Phys Lett A 253:358
Barbi M, Cocco S, Peyrard M, Ruffo S (1999) A twist opening model for DNA. J Biol Phys 24:97
Bates AD, Maxwell A (2009) DNA topology. Oxford University Press, Oxford
Calladine CR, Drew HR (1992) Understanding DNA. Academic Press, San Diego
Campa A, Giansanti A (1998) Experimental tests of the Peyrard-Bishop model applied to the melting of very short DNA chains. Phys Rev E 58:3585–3588
Chhetri KB, Dasgupta C, Maiti PB (2022) Diameter dependent melting and softening of dsDNA under cylindrical confinement. Front Chem 10:879746
Choi CH, Kalosakas G, Rasmussen KØ, Hiromura M, Bishop AR, Usheva A (2004) DNA dynamically directs its own transcription initiation. Nucl Acid Res 32:1584–1590
Cule D, Hwa T (1997) Denaturation of heterogeneous DNA. Phys Rev Lett 79:2375–2378
Dauxois T, Peyrard M, Bishop AR (1993) Entropy driven DNA denaturation. Phys Rev E 47:R44–R47
Depew RE, Wang JC (1975) Conformational fluctuations of DNA helix. Proc Natl Acad Sci USA 72:4275–4279
Drukker K, Wu G, Schatz GC (2001) Model simulations of DNA denaturation dynamics. J Chem Phys 114:579–590
Duguet M (1993) The helical repeat of DNA at high temperature. Nucl Acids Res 21:463–468
Englander SW, Kallenbach NR, Heeger AJ, Krumhansl JA, Litwin A (1980) Nature of the open state in long polynucleotide double helices: possibility of soliton excitations. Proc Natl Acad Sci USA 77:7222–7226
Feynman RP, Hibbs AR (1965) Quantum mechanics and path integrals. Mc Graw-Hill, New York
Gresham D, Dunham MJ, Botstein D (2008) Comparing whole genomes using DNA microarrays. Nat Rev 9:291
Guéron M, Leroy J-L (1985) Methods in enzymology 261. Academic Press, San Diego, USA
Hillebrand M, Kalosakas G, Bishop AR, Skokos Ch (2021) Bubble lifetimes in DNA gene promoters and their mutations affecting transcription. J Chem Phys 155:095101
Hillebrand M, Kalosakas G, Skokos Ch, Bishop AR (2020) Distributions of bubble lifetimes and bubble lengths in DNA. Phys Rev E 102:062114
Joshi H, Dwaraknath A, Maiti PK (2015) Structure, stability and elasticity of DNA nanotubes. Phys Chem Chem Phys 17:1424–1434
King M-C, Wilson AC (1975) Evolution at two levels in humans and chimpanzees. Science 188:107
Landau LD, Lifshitz EM (1980) Statistical physics. Pergamon Press, Oxford, UK
Landau LD, Lifshitz EM (2000) Quantum mechanics, 3rd edn. Butterworth-Heinemann, Oxford, UK
Le TT, Kim HD (2013) Measuring shape-dependent looping probability of DNA. Biophys J 104:2068–2076
Le TT, Kim HD (2014) Probing the elastic limit of DNA bending. Nucl Acids Res 42:10786–10794
Macedo DX, Guedes I, Albuquerque EL (2014) Thermal properties of a DNA denaturation with solvent interaction. Phys A 404:234–241
Majumdar SN (2005) Brownian functionals in Physics and Computer Science. Curr Sci 89:2076
Montgomery J, Wittwer CT, Palais R, Zhou L (2007) Simultaneous mutation scanning and genotyping by high-resolution DNA melting analysis. Nat Prot 2:59
Oliveira LM, Long AS, Brown T, Fox KR, Weber G (2020) Melting temperature measurement and mesoscopic evaluation of single, double and triple DNA mismatches. Chem Sci 11:8273–8287
Owczarzy R, You Y, Moreira BG, Manthey JA, Huang L, Behlke MA, Walder JA (2004) Effects of sodium ions on DNA duplex oligomers: improved predictions of melting temperatures. Biochemistry 43:3537–3554
Padinhateeri R, Menon GI (2013) Stretching and bending fluctuations of short DNA molecules. Biophys J 104:463–471
Peyrard M, Bishop AR (1989) Statistical mechanics of a nonlinear model for DNA denaturation. Phys Rev Lett 62:2755–2758
Peyrard M, Cuesta-López S, Angelov D (2009) Experimental and theoretical studies of sequence effects on the fluctuation and melting of short DNA molecules. J Phys: Condens Matter 21:034103
Peyrard M, Cuesta-López S, James G (2008) Modelling DNA at the mesoscale: a challenge for nonlinear science? Nonlinearity 21:T91–T100
Poland D, Scheraga H (1966) Phase transitions in one dimension and the Helix-Coil transition in polyamino acids. J Chem Phys 45:1456
Prohofsky EW (1988) Solitons hiding in DNA and their possible significance in RNA transcription. Phys Rev A 38:1538–1541
Romero-Enrique JM, de los Santos F, Muñoz MA (2010) Renormalisation group determination of the order of the DNA denaturation transition. EPL - Europhys Lett 89:40011
Saiki RK, Scharf S, Faloona F, Mullis KB, Horn GT, Erlich HA, Arnheim N (1975) Enzymatic amplification of P-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science 230:1350
SantaLucia J Jr, Hicks D (2004) The thermodynamics of DNA structural motifs. Annu Rev Biophys Biomol Struct 33:415
Scalapino DJ, Sears M, Ferrel RA (1972) Statistical mechanics of one-dimensional Ginzburg-Landau fields. Phys Rev B 6:3409
Schulman LS (1981) Techniques and applications of path integration. Wiley, New York
Shimada J, Yamakawa H (1984) Ring-closure probabilities for twisted Wormlike chains. Application to DNA. Macromolecules 17:689–698
Singh A, Modi T, Singh N (2016) Opening of DNA chain due to force applied on different locations. Phys Rev E 94:032410
Singh A, Singh N (2015) Effect of salt concentration on the stability of heterogeneous DNA. Phys A 419:328–334
Srivastava S, Singh N (2011) The probability analysis of opening of DNA. J Chem Phys 134:115102
Strick T, Allemand J-F, Croquette V, Bensimon D (2000) Twisting and stretching single DNA molecules. Prog Biophys Mol Biol 74:115–140
Sulaiman A, Zen FP, Alatas H, Handoko LT (2012) The thermal denaturation of the Peyrard-Bishop model with an external potential. Phys Scr 86:015802
Vafabakhsh R, Ha T (2012) Extreme bendability of DNA less than 100 base pairs long revealed by single-molecule cyclization. Science 337:1097–1101
van Eijck L, Merzel F, Rols S, Ollivier J, Forsyth VT, Johnson MR (2011) Direct determination of the base-pair force constant of DNA from the acoustic phonon dispersion of the double helix. Phys Rev Lett 107:088102
van Hove L (1950) Sur L’ intégrale de Configuration Pour Les Systèmes De Particules À Une Dimension. Physica 16:137–143
Wang JC (1979) Helical repeat of DNA in solution. Proc Natl Acad Sci USA 76:200–203
Wartell RM, Benight AS (1985) Thermal denaturation of DNA molecules: a comparison of theory with experiment. Phys Rep 126:67–107
Yakushevich LV (1994) Nonlinear DNA dynamics: hierarchy of the models. Physica D 79:77–86
Yamakawa H, Stockmayer WH (1972) Statistical mechanics of wormlike chains. II. Excluded volume effects. J Chem Phys 57:2843
Zhang F, Collins MA (1995) Model simulations of DNA dynamics. Phys Rev E 52:4217
Zhang YL, Zheng WM, Liu JX, Chen YZ (1997) Theory of DNA melting based on the Peyrard-Bishop model. Phys Rev E 56:7100–7115
Zoli M (1997) c-axis resistivity in high \(T_c\) superconductors. Phys Rev B 56:111
Zoli M (2004) Phonon thermodynamics versus electron-phonon models. Phys Rev B 70:184301
Zoli M (2005) Path integral of the two dimensional Su-Schrieffer-Heeger model. Phys Rev B 71:205111
Zoli M (2007) Finite size effects in bistable \(\phi ^4\) models. J Math Phys 48:012111
Zoli M (2009) Path integral method for DNA denaturation. Phys Rev E 79:041927
Zoli M (2010) Denaturation patterns in heterogeneous DNA. Phys Rev E 81:051910
Zoli M (2011a) Stacking interactions in denaturation of DNA fragments. Eur Phys J E 34:68
Zoli M (2011b) Thermodynamics of twisted DNA with solvent interaction. J Chem Phys 135:115101
Zoli M (2012) Anharmonic stacking in supercoiled DNA. J Phys: Condens Matter 24:195103
Zoli M (2013) Helix untwisting and bubble formation in circular DNA. J Chem Phys 138:205103
Zoli M (2014a) Twisting and bending stress in DNA minicircles. Soft Matter 10:4304–4311
Zoli M (2014b) Entropic penalties in circular DNA assembly. J Chem Phys 141:174112
Zoli M (2014c) Twist versus nonlinear stacking in short DNA molecules. J Theor Biol 354:95–104
Zoli M (2016a) Flexibility of short DNA helices under mechanical stretching. Phys Chem Chem Phys 18:17666
Zoli M (2016b) J- factors of short DNA molecules. J Chem Phys 144:214104
Zoli M (2017) Twist-stretch profiles of DNA chains. J Phys: Condens Matter 29:225101
Zoli M (2018a) Twisting short dsDNA with applied tension. Phys A 492:903–915
Zoli M (2018b) Short DNA persistence length in a mesoscopic helical model. EPL - Europhys Lett 123:68003
Zoli M (2018c) End-to-end distance and contour length distribution functions of DNA helices. J Chem Phys 148:214902
Zoli M (2019) DNA size in confined environments. Phys Chem Chem Phys 21:12566
Zoli M (2020a) First-passage probability: a test for DNA Hamiltonian parameters. Phys Chem Chem Phys 22:26901–26909
Zoli M (2020b) Stretching DNA in hard-wall potential channels. EPL - Europhys Lett 130:28002
Zoli M (2021) Base pair fluctuations in helical models for nucleic acids. J Chem Phys 154:194102
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zoli, M. Non-linear Hamiltonian models for DNA. Eur Biophys J 51, 431–447 (2022). https://doi.org/10.1007/s00249-022-01614-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00249-022-01614-z