Abstract
Fungi contribute to different important ecological processes, including decomposition of organic matter and nutrient cycling, but in the marine environment the main factors influencing their diversity and dynamics at the spatial and temporal levels are still largely unclear. In this study, we performed DNA metabarcoding on seawater sampled monthly over a year and a half in the Gulf of Trieste (northern Adriatic Sea), targeting the internal transcribed spacer (ITS) and the 18S rRNA gene regions. The fungal communities were diverse, very dynamic, and belonged predominantly to marine taxa. Samples could be clustered in two groups, mainly based on the high (> 30%) or low relative proportion of the ascomycetes Parengyodontium album, which emerged as a key taxon in this area. Dissolved and particulate organic C:N ratio played important roles in shaping the mycoplankton assemblages, suggesting that differently bioavailable organic matter pools may be utilized by different consortia. The proportion of fungal over total reads was 31% for ITS and 0.7% for 18S. ITS had the highest taxonomic resolution but low power to detect early divergent fungal lineages. Our results on composition, distribution, and environmental drivers extended our knowledge of the structure and function of the mycobiome of coastal waters.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In the marine environment, fungi have been detected in every explored habitat, from surface to deeper waters, from the coast to the open ocean, and from beaches to deep sediments [1, 2]. However, compared to their terrestrial counterparts, planktonic marine fungi, belonging to the mycoplankton, have been much less studied in terms of occurrence, biodiversity, dynamics, and contribution to ecosystem processes [3, 4]. Like terrestrial species, marine fungi are thought to contribute to organic matter degradation processes and nutrient cycling by acting as saprotrophic (i.e., decomposers) or parasitic organisms at different trophic levels [5]. For example, fungal zoospores efficiently transfer organic matter from large, otherwise inedible, phytoplankton cells to zooplankton, in a process termed mycoloop [6]. While mycoplankton are thought to have significant impacts on ecosystems, these organisms remain poorly understood [7].
More than 10,000 marine fungal species are estimated to live in the ocean, although less than 1900 have been formally described to date [8] (https://www.marinefungi.org/). Recently, after debated opinions [5], the scientific community has agreed on a common definition to classify a marine fungus, i.e., “it is repeatedly recovered from marine habitats because it is able to grow and/or sporulate in marine environments, it forms symbiotic relationships with other organisms, or it adapts and evolves at the genetic level or is metabolically active in the marine environment” [5]. While mycoplankton research has traditionally relied mainly on microscopic and culture-based approaches [5], the use of molecular tools is now overcoming the limitations of cultivation-based methods and revealing a large, previously unknown biodiversity [1, 7, 9]. These approaches, nonetheless, offer their own challenges. For example, passive propagule dispersion should be taken into account when assessing the role of fungi in marine habitats [10] when applying culture-independent, DNA-based approaches to estimate fungal diversity in near-shore environments.
Marine fungi have generally been overlooked in the context of global, ground-breaking programmes such as the Global Ocean Sampling expedition [11] and TARA Oceans [12], which have provided unprecedented resources for the genomics of marine prokaryotes and other eukaryotes [9]. One of the main limiting factors for DNA-based assessment of mycoplankton is the bias from "generic" eukaryotic primers (targeting the 18S rRNA gene [13]), which can lead to a lack of amplified fragments and consequently an underestimation of the proportion and diversity of fungi in biological communities [9, 14]. Therefore, the use of fungal-specific barcodes, such as those targeting the ribosomal nuclear gene internal transcribed spacer (ITS) [15] or the 18S rRNA gene [16], can provide greater insight into marine fungi biodiversity, although requiring ad hoc amplification, sequencing, and data analysis. In addition, mycoplankton can have patchy and highly variable spatiotemporal distribution patterns compared to bacterioplankton [9, 15, 17], making data analysis and interpretation more complex. Therefore, additional efforts are needed to explore the diversity and ecological role of fungi in marine habitats.
Here, we provide new insights into fungal communities in coastal environments, where mycoplankton are thought to play a particularly important role in organic matter cycling [4]. In this perspective, Long Term Ecological Research (LTER) stations represent a valuable resource and strategic advantage to assess patterns of mycoplankton diversity [15]. At the LTER C1 station (Gulf of Trieste, northern Adriatic Sea), seawater was sampled monthly over 1.5 years, and DNA metabarcoding targeting the ITS1 region was used to investigate mycoplankton composition, distribution, and key environmental factors influencing their temporal patterns, an effort directed to expand current knowledge of the structure and dynamics of coastal marine mycobiome. As a proof of concept, the fungal community was also investigated through 18S V4 rRNA gene metabarcoding to assess and compare the proportion and identity of the taxa identified with the two target regions.
Methods
Sample Collection and Environmental Data
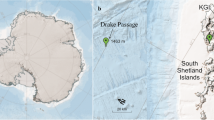
The sampling site was the LTER monitoring station C1 (45° 42′ 2″ N, 13° 42′ 36″ E; Fig. 1) located in the Gulf of Trieste (northern Adriatic Sea). Seawater samples were collected monthly between October 2018 and April 2020 at the surface (~ 1 m depth, S) and at the bottom (15 m depth, B) of the water column using 5-L Niskin bottles. Samples were filtered through 0.2 μm PES membrane filters (PALL Laboratory) until clogging (1–3 L) and stored at −80 °C until further processing. In the frame of the LTER monitoring at station C1, contextually to DNA sampling, a set of biogeochemical parameters was determined. Temperature and salinity were measured by means of a multiparametric probe (SBE 19plus SEACAT). The concentrations of chlorophyll a (Chla), dissolved organic carbon (DOC), particulate organic carbon (POC), dissolved organic nitrogen (DON), and total particulate nitrogen (TPN) were determined according to standard procedures [18, 19] (Supplementary results and Fig. S1).
C1 sampling site location in the northern Adriatic Sea. Maps designed with Ocean Data View (https://odv.awi.de)
DNA Extraction and Amplicon Sequencing
DNA was extracted using the DNeasy PowerWater Kit (Qiagen) with some modifications (two additional vortexing steps for 2 min at the maximum speed, each one preceded by an incubation at 70 °C for 5 min) [20].
For amplicon sequencing, the nuclear ribosomal internal transcribed spacer 1 (ITS1) region was amplified in all samples (Surface and Bottom, for a total of 38 samples) using the primers ITS1-F [21] and ITS2-R [22]. Libraries were prepared following the Illumina Sequencing Library Preparation protocol (with 10 µL of DNA and 35 PCR cycles in the amplicon PCR) and ran on an Illumina MiSeq System for a read length of 2 × 250 bp at Cogentech (Consortium for Genomic Technologies c/o IFOM-IEO Campus, Milano, Italy).
The V4 region of the 18S rRNA (18S V4) gene was amplified only in the surface samples (for a total of 19 samples) using the "generic" eukaryotic primers TAReuk454FWD1 [13] and TAReukREV3_modified [23, 24]. Libraries were prepared following the Illumina Sequencing Library Preparation protocol and ran on an Illumina MiSeq System for a read length of 2 × 250 bp BMR genomics (Padova, Italy).
Bioinformatic Pipelines
For the ITS1 barcode, the PIPITS v. 3.0 pipeline [25] was used at default parameters for reads merging and quality filtering. ITS1 region was extracted with ITSx [25]. Then, operational taxonomic unit (OTU) clustering at 97% similarity and chimera removal were performed with VSEARCH v. 2.23.0 [25]. OTUs with a frequency < 10 were removed. Alpha-diversity metrics were estimated after samples were rarefied.
Taxonomy was assigned to OTUs within QIIME2 (v. 2023.5) [26] using the sklearn naïve Bayes taxonomy classifier against the UNITE reference database (v. 9) [27]. BLASTN [28] was also used as an additional check, aligning the representative sequences against the nucleotide collection (BLAST + 2.14.1).
Finally, only sequences belonging to fungi (fungi—Kingdom Mycetae—and fungal-like organisms—Oomycetes, Hyphochytriomycetes, Labyrinthulomycetes in Kingdom Straminipila—[29]) were retained in the final dataset. A "core" mycobiome was identified by selecting the genera present in more than 95% of the samples. To assess similarity patterns of fungal communities, a hierarchical clustering of the OTU table (Ward.D2 method) was built using the function hclust in the R (v. 4.0.3) [30] package stats (v. 4.3.0). A permutational multivariate analysis of variance (PERMANOVA) with 4999 permutations was computed on the normalized OTU table using the function adonis in the R (v. 4.0.3) [30] package vegan (v. 2.6.4) [31]. To investigate the fungal community environmental drivers, a distance-based redundancy analysis (dbRDA) was performed on Bray-Curtis dissimilarity matrices on the OTU table using the function capscale in the R (v. 4.0.3) [30] package vegan (v. 2.6.4) [31]. The environmental variables were first screened for collinearity [32], and the model and variable significance were tested with ANOVA and 4999 permutations. Marine genera were assessed following comprehensive works and reviews based on both traditional and molecular methods [2, 8, 10, 33]. Marine genera were assessed considering only taxa that presented > 0.01% of average relative abundance (considered non-rare taxa [34]). FUNGuild [35] was used to predict marine genera trophic and growth modes, functional guilds, and habitat type.
For the 18S V4 barcode, analyses were performed with QIIME2 (v. 2023.5) [26] using DADA2 for denoising. Amplicon sequence variants (ASVs) with a frequency < 2 were removed. Taxonomy was assigned to ASVs using the sklearn naïve Bayes taxonomy classifier against the SILVA 99% reference database (v. 138.1) [36], and only ASVs belonging to Fungi were retained.
Results
Fungal Community
The ITS1 amplicon sequencing of the 38 samples generated 5,836,517 raw reads. After cleaning, ITS1 extraction, and chimera removal, 4,528,841 reads were retained with an average of 119,180 ± 59,972 per sample and a total of 6885 OTUs with an average of 1170 ± 577 per sample. Sample Dec19_B was removed as 92.2% of its reads belonged to a cnidaria. After removing the non-fungal sequences (all of them belonging to the Myceteae), 1,396,478 reads (31% of the total dataset) were retained. The total number of OTUs was 2017 with an average of 246 (± 196) per sample. Shannon’s diversity index was, on average, 3.5 (± 1.7), while Pielou’s evenness was 0.50 (± 0.17).
The taxonomy of the fungal OTUs in each sample, presented as Krona charts [37], is available at https://github.com/ElisaBanchi/Fungi_ITS. Six phyla were detected, with most reads assigned to Ascomycota (76.5 ± 25.3% on average) (Fig. 2). Ascomycota was generally the dominant phylum (in 34 out of 38 samples; Fig. 2), while Basidiomycota prevailed in a surface winter water sample (Feb20_S; Fig. 2) and Chytridiomycota in early spring (Fig. 2).
A total of 772 genera (of which 653 were identified with a generic name; Table S1) were detected, the most abundant being Parengyodontium (a monospecific genus with the only known species P. album, 37.2 ± 33.0%; Fig. 2). Only 14 genera reached an average abundance > 0.5% (Fig. 2), while 505 genera could be considered rare (< 0.01%; Table S1).
We identified six genera as representatives of the core mycobiome in the entire data set, i.e., Aspergillus, Cladosporium, Exophiala, Malassezia, Parengyodontium, and Penicillium.
Due to the highly dynamic taxonomic composition across the dataset (Fig. 2), we performed hierarchical clustering to highlight the similarity among samples. Samples fell into two well-distinct clusters (Fig. S2), with Cluster 1 comprising all samples in which the relative proportion of P. album was greater than 30% (Fig. S2). PERMANOVA showed that the separation of the clusters explained 82% of the variance (p < 0.001).
The dbRDA (Fig. 3) showed that DOC/DON and TOC/TPN ratios had significant roles (p < 0.001 and p < 0.05 respectively) in shaping the fungal communities and in differentiating samples in terms of diverse proportion of P. album.
Distance-based redundancy analysis (dbRDA) based on Bray-Curtis dissimilarity in community composition. T = temperature; S = salinity; DOC = dissolved organic carbon; POC = particulate organic carbon; DON = dissolved organic nitrogen; TPN = total particulate nitrogen. Asterisks indicate significant environmental variables
Multivariate analysis did not highlight significant differences between surface and bottom samples in terms of taxonomic composition and environmental drivers.
Marine Genera
Among the non-rare 227 defined genera (Table S1), 47 were defined as marine, representing 82% of the total dataset in terms of relative abundance. The corresponding OTUs were 309 with an average of 69 ± 33 per sample, corresponding to 72.6 ± 25.8% of the reads on average per sample (Fig. 4).
The marine genera belonged to three phyla: Ascomycota, Basidiomycota, and Basidiobolomycota and included all "core" as well as all the most abundant genera (average > 0.5%) except for Knufia and Peniophora (Fig. 2). The dbRDA (Fig. S3) performed on this subset highlighted DOC/DON and TOC/TPN ratios as significant drivers (p < 0.001 and p < 0.05, respectively) of the marine fungal communities and in differentiating samples in terms of different proportion of P. album.
18S V4 Analysis
For 18S V4, a total of 3,790,777 raw reads were generated for the 19 samples. After primer removal, denoising, and chimera deletion, 2,873,322 reads were retained, with an average of 151,128 ± 50,679 reads and 740 ± 278 ASVs per sample for a total of 7177 ASVs. Most reads belonged to the phyla Dinoflagellata (23.6%), while the proportion of fungal taxa (Kingdom Fungi) was 0.7% (20,906 on 2,873,322 reads and 209 on 7177 ASVs). In only four samples (Mar19_S, Jul19_S, Nov18_S, Oct19_S, Feb19_S), the number of fungal reads was higher than 1000. Unlike for ITS1, reads belonging to Labyrinthulomycetes (Straminipila, 0.6% of the dataset) were detected.
The taxonomy of the samples, presented as interactive Krona charts [37], is available at https://github.com/ElisaBanchi/Fungi_ITS. Four phyla (Ascomycota, Basidiomycota, Chytridiomycota, and Rozellomycota) were detected (Fig. S4). The number of genera detected amounted to 47 (of which 41 were identified with a generic name). The most abundant (average > 0.5%; Fig. S4) genera were Cystobasidium, Gjaerumia, Kondoa, Malassezia, Metschnikowia, Nowakowskiella, Paramicrosporidium, Rhizophydium, Rhodotorula, and Sakaguchia.
Of the 41 genera identified with 18S V4, 31 were in common with ITS1 (considering only surface samples) (Table S2). Since 18S V4 had a low proportion of fungal reads, the number of taxa detected in each sample was correspondingly low. For example, the number of genera per sample averaged 4.5 ± 4 for 18S V4, while it was 144 ± 88 for ITS1. The different proportion and resolution of the fungal reads obtained with the two barcode genes is reflected in the different taxonomic composition of the same sample. An example of this is shown in Fig. 5 for Jan19_S. Here, the number of OTUs/ASVs was 10 for 18S V4 and 141 for ITS1, of which 4 and 122, respectively, could be assigned at the genus level. In addition to the relative abundances, presence/absence patterns were different: Chytridiomycota, for example, were found with 18S V4 but not with ITS1, while the contrary was true for Rozellomycota.
Discussion
A Diverse and Dynamic Mycobiome Shaped by Organic Matter Quality
Overall, the fungal community detected at the LTER station C1 was in line with results from recent DNA metabarcoding surveys in coastal and open waters [7, 15, 17, 29, 38, 39]. In fact, we detected a prevalence of Dikaryotic fungi, Ascomycota, and, in minor proportion, Basidiomycota, in surface and bottom samples (Fig. 2). Chytridiomycota, instead, were abundant in a few samples only. While we could assign most of the detected Ascomycota and Basidiomycota to the genus level, Chytridiomycota showed a lower taxonomic resolution. This also applies to the lower taxonomic levels, with the most common genera such as Aspergillus, Aureobasidium, Candida, Cladosporium, Cryptococcus, Malassezia, Penicillium, Parengyodontium, and Rhodotorula being among the taxa most frequently detected in marine ecosystems [2, 5, 15].
The fungal communities we detected included both yeasts (single-celled: Exophiala, Malassezia, Metschnikowia) and filamentous forms (hyphal: Acremonium, Aspergillus, Cladosporium, Parengyodontium, Penicillium), reflecting the complexity of this planktonic fraction. Moreover, from an “amplicon sequencing” point of view, the detection of organisms (or portions of them) with mycelial (filamentous, multicellular), unicellular (yeasts), and possibly multinucleate forms, as well as the number, size, and type of spores produced, could represent a bias in different steps of the workflow, including the relative proportion of reads.
A common feature of mycoplankton is its high dynamicity at both the spatial and temporal levels [2, 17], and with respect to other planktonic organisms (e.g., bacterioplankton), it is generally characterized by a patchier distribution, largely dependent on the quality and availability of organic carbon [40]. In fact, both yeast and filamentous fungi can be attached to large particles and can colonize microzones where there is a large availability of organic matter [29, 38]. Moreover, a similar situation can be hypothesized to happen in seawater as described for freshwater streams, where, based on the availability of substrates, fungal mycelia and conidia can show boom-bust cycles [41]. Chrismas et al. [15], in the longest time-series on mycoplankton (17 years), recovered different recurrence patterns: annually (e.g., Metschnikowia), persistent (e.g., Cladosporium, Symmetrospora), occasional (e.g., Rhodotorula, Parengyodontium), or random (e.g., Penicillium). Also, in our survey, the compositional patterns were rather variable and patchy along the sampling period at both depths. Indeed, none of the taxa was steadily present (Fig. 2), rather showing episodic peaks decoupled from the hydrological or biological features of the study area (Fig. 3), differently from what was described for bacterioplankton [42] and phytoplankton [43]. For this reason, we used a hierarchical clustering approach to highlight similarity among samples.
From this analysis, Parengyodontium album emerged as a key taxon of the northern Adriatic mycoplankton, as well as the most abundant (37.2 ± 33.0%, Fig. 2) and part of the core mycobiome. P. album is a filamentous, chemoorganotrophic species often detected in seawater [15, 39]. It has also been found in sediments and associated with sponges, corals, and crustaceans and regarded as a potential pathogen of marine animals [44]; it is also considered an emerging opportunistic human pathogen due to its proteolytic and keratinolytic activities [45]. Other abundant and part of the core mycobiome taxa of our C1 site such as Aspergillus, Cladosporium, Exophiala, and Penicillium (Fig. 2), together with other fungi present in lower abundance such as Rhodotorula, Mortierella, and Trichoderma, are known to be able to degrade recalcitrant and complex molecules including oil, hydrocarbons, and lignin [1, 46]. Cladosporium is a cosmopolitan fungal genus commonly detected in both terrestrial [47] and aquatic environments, with halo- and osmotolerant species adapted to marine environments [29]. Moreover, Cunliffe et al. [48] using DNA stable-isotope probing showed that well-represented marine strains of Cladosporium can assimilate algal-derived POC. The utilization of these compounds highlighted a saprotrophic functional role of these taxa in processing algal polysaccharides and, as they can potentially be eaten by zooplankton, a link for C transfer to higher trophic levels [48].
In marine ecosystems, in analogy with terrestrial processes, fungi can occur mainly as saprotrophs or symbionts, occupying a wide range of ecological niches [5]. Saprotrophic fungi, especially Dikarya, can secrete (exo-)hydrolytic enzymes to process labile as well as refractory organic molecules [38, 40]. These include recalcitrant materials (with high C:N ratios) that are more difficult for bacterioplankton to utilize, suggesting a complementary role for these two fractions [17]. Fungi can play an important role in marine food webs by building complex biotrophic interactions with phytoplankton and zooplankton (such as parasitism, predation, grazing, and pathogenicity) and can shape carbon fluxes within planktonic food webs via the mycoloop [6, 38, 40]. Besides the conceptual importance of mycoplankton, the main drivers of its compositional and diversity patterns, as well as its temporal and spatial dynamics, remain largely unclear [7, 17]. This is primarily due to the limited number of studies that have investigated these organisms, which are scattered both temporally and spatially (e.g., coastal vs open ocean) and therefore span different ecosystems and environmental gradients. In addition, the different barcode targets and taxonomic resolutions gained or considered make it difficult to integrate and compare the results of the different studies. In general, fungal communities are the result of the interplay of multiple environmental factors [17]. Breyer and Baltar [7] in their meta-analysis listed potential influencing drivers: (i) abiotic factors (such as temperature, depth, pH, salinity), (ii) the availability of major elements (C, N, P), and (iii) biotic factors (abundance and composition of phytoplankton and zooplankton).
In this study, no consistent role of chlorophyll a concentration was observed (Fig. 3), evidencing that the relationship between fungi and microalgae is mainly sporadic. On the other hand, dissolved (DOC/DON) and particulate (TOC/TPN) organic carbon and nitrogen ratios were significant drivers of the fungal communities (Fig. 3). In another study in the Mediterranean (Ligurian Sea), Celussi et al. [20] hypothesized that a higher presence of Fungi in samples with a higher C:N ratio could explain the observed higher rates of organic matter degradation. Similarly, Bochdansky et al. [49] found a high fungi-to-prokaryotes ratio, in terms of biomass, associated with deep-sea marine snow, rich in refractory organic compounds [50].
Noteworthy in our dataset, different mycoplankton assemblages were associated with either dissolved or particulate C:N ratio. For instance, in the samples characterized by higher dissolved C:N ratio, the yeast-forming genera Kondoa and Malassezia were more abundant, while the filamentous and cosmopolitan Cladosporium and Alternaria genera were more present in samples with higher particulate C:N ratio, suggesting that different fungal consortia may rely on these different fractions of recalcitrant organic matter. While the role of marine fungi in the processing of dissolved refractory compounds is difficult to quantify, there is evidence of their capability to degrade dissolved humic compounds (high C:N) through multiple degradation pathways [3]. Taken together, these findings strengthen the idea that in temperate marine waters, mycoplankton may play a complementary role alongside bacterioplankton in organic matter cycling, possibly breaking down complex organic compounds and thus increasing their bioavailability, boosting the microbial loop.
The Coastal Environment as a Sink for Terrestrial Fungi
Although this work focuses mainly on marine taxa, the identification and exploration of the terrestrial fraction are interesting and allow for a more comprehensive assessment of the biodiversity of the studied area, the influence of land use, and the evaluation of land-sea exchanges (e.g., input of organic matter and nutrients).
The occurrence of terrestrial fungi (Dikarya) in coastal and oceanic habitats has been linked to their reproductive success and airborne dispersal. Their spores can be transported over long distances and at high speed [33]; therefore, fungi living in woods or rocks can easily be transported in water. In this respect, coastal waters act as a sink for land-derived fungi [51], increasing mycobiome diversity. For example, Chrismas et al. [15] detected different genera associated with forests as well as a lichen-forming genus (Lichina) present along the coast near their sampling site. In our dataset, the most abundant non-marine genera were Knufia and Peniophora. Knufia is an oligotrophic and halotolerant, rock-inhabiting Ascomycota, a persistent dweller of natural and anthropogenic extreme habitats [52]. Even if the presence of fungi of terrestrial origin in seawater should be considered random, which is also related to higher or lower dispersal capabilities, we cannot exclude the possibility that halophilic taxa could survive in this environment. Peniophora, a filamentous Basidiomycota, is a widespread genus that grows on decaying wood and plant decay material [53]: it is likely that the amplified reads derived from mycelia on wood and truck pieces have entered the sea either transported by winds, rivers, floods, or tides.
Taxonomic Resolution and Detection of Early Divergent Fungi: ITS and 18S
The choice of a target region for DNA metabarcoding studies is a critical step as it influences the amplification procedures (e.g., amplicon length, number of cell/copies) as well as the bioinformatic pipelines and taxonomic assignment (e.g., reference databases). For fungal communities, a consensus has been found on the choice of internal transcribed spacer (ITS) [54], with studies targeting either subregions 1 (ITS1) or 2 (ITS2) [7] or even aiming at sequencing the whole ITS region through the latest third-generation sequencing technologies (e.g., Oxford Nanopore, Pacific Biosciences).
In this study, we identified different proportions of fungal reads obtained with the two barcodes (31% for ITS1 and 0.7% for 18S V4), resulting in a tenfold difference in the number of identified genera (Fig. 5, Table S2). If the 18S rRNA gene has lower variation and thus lower taxonomic accuracy and resolution than the ITS region [14], which also emerged in our study (e.g., for Hypocreales), the use of the ITS as a marker may introduce biases and lack the detection of some early-diverging lineages. For example, in 18S V4, we found that the proportion of Chytridiomycota was higher compared to ITS1, but still largely undefined at the lower taxonomic levels. However, some genera were identified, including the marine Olpidium (comprising mainly obligate endoparasites of algae, plants, fungi, and animals [55]) and Rhizophydium (comprising diatom parasites [56]). The assignment of Chytridiomycota reads to lower taxonomic levels is challenging due to the lack of reference sequences in public databases including UNITE [27] and to their high genetic divergence [56]. As a result, the contribution of this marine basal group to the transfer of organic matter and nutrients within food webs is likely underestimated [5]. Moreover, it is worth noticing that the pattern of Chytridiomycota abundance, which commonly depends on host-parasite biotic interactions with phytoplankton, is not always evident [57]. Indeed, we could not establish a clear relationship between chytrids and phytoplankton, possibly because the Chytridiomycota we detected could be associated with saprotrophic or parasitic organisms besides phytoplankton, as already suggested by Banos et al. [38]. We could also speculate that, if an event (e.g., a diatom bloom) occurred, it was too short to be captured by the sampling frequency we used for this study.
Overall, our study confirmed that ITS has a high taxonomic resolution within mycoplankton, but further improvement of reference databases is needed to better identify marine taxa, especially Chytridiomycota. If 18S data obtained with universal eukaryotic primers are available, screening of fungal reads may provide complementary information on early divergent fungal lineages. However, we suggest approaching these data in terms of presence/absence rather than in terms of relative abundance, to exploit the detection power of the barcode while at the same time partially accounting for the biases due to the low number of fungal reads.
Conclusion
This is, to the best of our knowledge, the first study targeting mycoplankton in the Adriatic Sea. The fungal communities in the C1 LTER coastal site were very diverse and dynamic, with Dikarya and sporadically Chytridiomycota as prevalent taxa. The quality and availability of organic matter likely play an important role in the distribution of fungi, and Parengyodontium album was identified as a key taxon in the mycoplankton of the northern Adriatic, but further efforts are needed to clarify its functional role in the coastal environment.
New insights into the characterization of mycoplankton could be gained by using long-reads DNA metabarcoding covering the 18S-ITS-28S region in conjunction with the isolation and cultivation of marine fungal strains potentially belonging to representative species and to early divergent lineages.
Improving the spatial and taxonomic resolution of the marine fungal community will help to clarify the role of mycoplankton in marine food webs and biogeochemical cycles and to develop an integrated picture of the structure and functions of the coastal environment.
Data Availability
The nucleotide sequence data reported are available in the Sequence Reads Archive (SRA) at NCBI under the accession numbers PRJNA1061483.
References
Amend A, Burgaud G, Cunliffe M et al (2019) Fungi in the marine environment: open questions and unsolved problems. MBio10:e01189-18. https://doi.org/10.1128/mBio.01189-18
Grossart HP, Van den Wyngaert S, Kagami M et al (2019) Fungi in aquatic ecosystems. Nat Rev Microbiol 17:339–354. https://doi.org/10.1038/s41579-019-0175-8
Grossart HP, Rojas-Jimenez K (2016) Aquatic fungi: targeting the forgotten in microbial ecology. COMICR 31:140–145. https://doi.org/10.1016/j.mib.2016.03.016
Wang Y, Sen K, He Y et al (2019) Impact of environmental gradients on the abundance and diversity of planktonic fungi across coastal habitats of contrasting trophic status. STOTEN 683:822–833. https://doi.org/10.1016/j.scitotenv.2019.05.204
Burgaud G, Edgcomb VP, Hassett BT et al (2022) Marine fungi. In: Stal LJ and Cretoiu MS (ed) The marine microbiome. The microbiomes of humans, animals, plants, and the environment. Berlin, pp 243–95
Kagami M, Miki T, Takimoto G (2014) Mycoloop: chytrids in aquatic food webs. Front Microbiol 5:166. https://doi.org/10.3389/fmicb.2014.00166
Breyer E, Baltar F (2023) The largely neglected ecological role of oceanic pelagic fungi. Trends Ecol Evol 38:870–888. https://doi.org/10.1016/j.tree.2023.05.002
Jones EBG, Pang KL, Abdel-Wahab MA et al (2019) An online resource for marine fungi. Fungal Divers 96:347–433. https://doi.org/10.1007/s13225-019-00426-5
Gladfelter AS, James TY, Amend AS (2019) Marine fungi. Curr Biol 29:191–195. https://doi.org/10.1016/j.cub.2019.02.009
Jones EG, Suetrong S, Sakayaroj J et al (2015) Classification of marine ascomycota, basidiomycota, blastocladiomycota and chytridiomycota. Fungal Divers 73:1–72. https://doi.org/10.1007/s13225-015-0339-4
Rusch DB, Halpern AL, Sutton G et al (2007) The Sorcerer II global ocean sampling expedition: northwest Atlantic through eastern tropical Pacific. PLoS Biol 5:e77. https://doi.org/10.1371/journal.pbio.0050077
Sunagawa S, Acinas SG, Bork P et al (2020) Tara Oceans: towards global ocean ecosystems biology. Nat Rev Microbiol 18:428–445. https://doi.org/10.1038/s41579-020-0364-5
Stoeck T, Bass D, Nebel M et al (2010) Multiple marker parallel tag environmental DNA sequencing reveals a highly complex eukaryotic community in marine anoxic water. Mol Ecol 19:21–31. https://doi.org/10.1111/j.1365-294X.2009.04480.x
Richards TA, Leonard G, Mahé F et al (2015) Molecular diversity and distribution of marine fungi across 130 European environmental samples. Proc Royal Soc B 282:20152243. https://doi.org/10.1098/rspb.2015.2243
Chrismas N, Allen R, Allen MJ et al (2023) A 17-year time-series of fungal environmental DNA from a coastal marine ecosystem reveals long-term seasonal-scale and inter-annual diversity patterns. Proc Royal Soc B 290:2022–2129. https://doi.org/10.1098/rspb.2022.2129
Banos S, Lentendu G, Kopf A et al (2018) A comprehensive fungi-specific 18S rRNA gene sequence primer toolkit suited for diverse research issues and sequencing platforms. BMC Microbiol 18:1–15. https://doi.org/10.1186/s12866-018-1331-4
Duan Y, Xie N, Wang Z et al (2021) Patchy distributions and distinct niche partitioning of mycoplankton populations across a nearshore to open ocean gradient. Microbiol Spectr 9:e01470–e01421. https://doi.org/10.1128/Spectrum.01470-21
Manna V, De Vittor C, Giani M et al (2021) Long-term patterns and drivers of microbial organic matter utilization in the northernmost basin of the Mediterranean Sea. Mar Environ Res 164:105245. https://doi.org/10.1016/j.marenvres.2020.105245
Schroeder K, Kovačević V, Civitarese G et al (2024) Seawater physics and chemistry along the Med-SHIP transects in the Mediterranean Sea in 2016. Sci Data 11:52. https://doi.org/10.1038/s41597-023-02835-3
Celussi M, Quero GM, Zoccarato L et al (2018) Planktonic prokaryote and protist communities in a submarine canyon system in the Ligurian Sea (NW Mediterranean). Prog Oceanog 168:210–221. https://doi.org/10.1016/j.pocean.2018.10.002
Gardes M, Bruns TD (1993) ITS primers with enhanced specificity for basidiomycetes-application to the identification of mycorrhizae and rusts. Mol Ecol 2:113–118. https://doi.org/10.1111/j.1365-294x.1993.tb00005.x
White TJ, Bruns T, Lee S et al (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Michael A, Innis M, Gelfand DH, Sninsky JJ et al (eds) PCR protocols a guide to methods and applications. Academic, San Diego, pp 315–322
Piredda R, Tomasino MP, D’erchia AM et al (2017) Diversity and temporal patterns of planktonic protist assemblages at a Mediterranean Long Term Ecological Research site. FEMS Microbiol Ecol 93:fiw200. https://doi.org/10.1093/femsec/fiw200
Kettner MT, Oberbeckmann S, Labrenz M et al (2019) The eukaryotic life on microplastics in brackish ecosystems. Front Microbiol 10:538. https://doi.org/10.3389/fmicb.2019.00538
Gweon HS, Oliver A, Taylor J et al (2015) PIPITS: an automated pipeline for analyses of fungal internal transcribed spacer sequences from the Illumina sequencing platform. Methods ecol evol 6:973–980. https://doi.org/10.1111/2041-210X.12399
Bolyen E, Rideout JR, Dillon MR et al (2019) Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 37:852–857. https://doi.org/10.1038/s41587-019-0209-9
Abarenkov K, Zirk A, Piirmann T et al UNITE QIIME release for Fungi. 2022 UNITE Community. https://doi.org/10.15156/BIO/2483915
Altschul SF, Madden TL, Schäffer A et al (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. https://doi.org/10.1093/nar/25.17.3389
Raghukumar S (2001) Fungi in coastal and oceanic marine ecosystems. Springer, New York
R Core Team (2021) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/
Oksanen J, Blanchet FG, Kindt R et al (2013) Community ecology package. R package version 2.5-4 2:321–326
Zuur AF, Ieno EN, Elphick CS (2010) A protocol for data exploration to avoid common statistical problems. Methods Ecol Evol 1:3–14. https://doi.org/10.1111/j.2041-210X.2009.00001.x
Hassett BT, Vonnahme TR, Peng X et al (2019) Global diversity and geography of planktonic marine fungi. Bot Mar 63:121–139. https://doi.org/10.1515/bot-2018-0113
Gobet A, Böer S, Huse SM et al (2012) Diversity and dynamics of rare and of resident bacterial populations in coastal sands. ISME J 6:542–553. https://doi.org/10.1038/ismej.2011.132
Nguyen NH, Song Z, Bates ST et al (2016) FUNGuild: an open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol 20:241–248. https://doi.org/10.1016/j.funeco.2015.06.006
Quast C, Pruesse E, Yilmaz P et al (2012) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acid Res 41:590–596. https://doi.org/10.1093/nar/gks1219
Ondov BD, Bergman NH, Phillippy AM (2011) Interactive metagenomic visualization in a web browser. BMC Bioinform 12:1–10. https://doi.org/10.1186/1471-2105-12-385
Banos S, Gysi DM, Richter-Heitmann T et al (2020) Seasonal dynamics of pelagic mycoplanktonic communities: interplay of taxon abundance, temporal occurrence, and biotic interactions. Front Microbiol 11:1305. https://doi.org/10.3389/fmicb.2020.01305
Huang J, Zhao H, Yang S et al (2022) Mycoplanktonic community structure and their roles in monitoring environmental changes in a subtropical estuary in the Beibu gulf. J Mar Sci Eng 10:1940. https://doi.org/10.3390/jmse10121940
Wang G, Wang X, Liu X et al (2012) Diversity and biogeochemical function of planktonic fungi in the ocean. Prog Mol Subcell Biol 53:71–88. https://doi.org/10.1007/978-3-642-23342-5_4
Bärlocher F, Boddy L (2016) Aquatic fungal ecology–how does it differ from terrestrial? Fungal Ecol 19:5–13. https://doi.org/10.1016/j.funeco.2015.09.001
Celussi M, Manna V, Banchi E et al (2024) Annual recurrence of prokaryotic climax communities in shallow waters of the North Mediterranean. Environ Microbiol 26:e16595. https://doi.org/10.1111/1462-2920.16595
Cerino F, Fornasaro D, Kralj M et al (2019) Phytoplankton temporal dynamics in the coastal waters of the north-eastern Adriatic Sea (Mediterranean Sea) from 2010 to 2017. Nat Conserv 34:343–372. https://doi.org/10.3897/natureconservation.34.30720
Shaumi A, Cheang UC, Yang CY et al (2021) Culturable fungi associated with the marine shallow-water hydrothermal vent crab Xenograpsus testudinatus at Kueishan Island, Taiwan. Bot Mar 64:289–300. https://doi.org/10.1515/bot-2021-0034
Tsang CC, Chan JF, Pong WM et al (2016) Cutaneous hyalohyphomycosis due to Parengyodontium album gen. et comb. nov. Med Mycol 54:699–713. https://doi.org/10.1093/mmy/myw025
Hara E, Uchiyama H (2012) Degradation of petroleum pollutant materials by fungi. In: Goltapeh E, Danesh Y, Varma A (eds) Fungi as Bioremediators. Springer, Berlin, pp 117–133
Tordoni E, Ametrano CG, Banchi E et al (2021) Integrated eDNA metabarcoding and morphological analyses assess spatio-temporal patterns of airborne fungal spores. Ecol Ind 121:107032. https://doi.org/10.1016/j.ecolind.2020.107032
Cunliffe M, Hollingsworth A, Bain C et al (2017) Algal polysaccharide utilisation by saprotrophic planktonic marine fungi. Fungal Ecol 30:135–138. https://doi.org/10.1016/j.funeco.2017.08.009
Bochdansky AB, Clouse MA, Herndl GJ (2017) Eukaryotic microbes, principally fungi and labyrinthulomycetes, dominate biomass on bathypelagic marine snow. ISME 11:362–373. https://doi.org/10.1038/ismej.2016.113
Baltar F, Arístegui J, Sintes E et al (2010) Significance of non-sinking particulate organic carbon and dark CO2 fixation to heterotrophic carbon demand in the mesopelagic northeast Atlantic. Geophys Res Lett 37. https://doi.org/10.1029/2010GL043105
Vass M, Eriksson K, Carlsson-Graner U et al (2022) Co-occurrences enhance our understanding of aquatic fungal metacommunity assembly and reveal potential host–parasite interactions. FEMS Microbiol Ecol 98:fiac120. https://doi.org/10.1093/femsec/fiac120
Sterflinger K (2006) Black yeasts and meristematic fungi: ecology, diversity and identification. In: Péter G, Rosa C (eds) Biodiversity and Ecophysiology of yeasts. Springer, Berlin, pp 501–514
Cavanna C, Pagella F, Esposto MC et al (2019) Human infections due to Schizophyllum commune: case report and review of the literature. J Mycol Med 29:365–371. https://doi.org/10.1016/j.mycmed.2019.100897
Xu J (2016) Fungal DNA barcoding. Genome 59:913–932. https://doi.org/10.1139/gen-2016-0046
Chang Y, Rochon DA, Sekimoto S et al (2021) Genome-scale phylogenetic analyses confirm Olpidium as the closest living zoosporic fungus to the non-flagellated, terrestrial fungi. Sci rep 11:3217. https://doi.org/10.1038/s41598-021-82607-4
Frenken T, Alacid E, Berger SA et al (2017) Integrating chytrid fungal parasites into plankton ecology: research gaps and needs. Environ Microbiol 19:3802–3822. https://doi.org/10.1111/1462-2920.13827
Reñé A, Timoneda N, Sarno D et al (2023) Vertical and temporal distribution of chytrids infecting diatoms in the Gulf of Naples (Italy, Mediterranean Sea). Mar Ecol 44:e12726. https://doi.org/10.1111/maec.12726
Acknowledgements
The authors would like to thank M. Bazzaro (OGS), M. Kralj (OGS), and F. Relitti (OGS) for chemical analyses, V. Fonti (OGS) for DNA extraction, and G. Zazo (OGS) for CTD casts and data processing. The comments of two anonymous reviewers helped improve the original version of the manuscript.
Funding
Open access funding provided by Istituto Nazionale di Oceanografia e di Geofisica Sperimentale within the CRUI-CARE Agreement. This work was supported by the Interreg MED Strategic Project SHAREMED, co-financed by the European Regional Development Fund under the Funding Programme Interreg MED 2014–2020 (https://sharemed.interreg-med.eu/) and benefited from CINECA ISCRA C resources (HP10CPNSKZ; PI: E.B.). E.B., M.C., and V.M. are partially supported by the project “National Biodiversity Future Center - NBFC” funded under the National Recovery and Resilience Plan (NRRP), Mission 4 Component 2 Investment 1.4 - Call for tender No. 3138 of 16 December 2021, rectified by Decree n.3175 of 18 December 2021 of Italian Ministry of University and Research funded by the European Union – NextGenerationEU; Project code CN_00000033, Concession Decree No. 1034 of 17 June 2022 adopted by the Italian Ministry of University and Research, CUP F83B22000050001.
Author information
Authors and Affiliations
Contributions
E.B.: conceptualization, methodology, formal analysis, and writing—original draft, review, and editing. V.M.: writing—review and editing. L.M.: writing—review and editing. M.C.: conceptualization, resources, and writing—review and editing.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Banchi, E., Manna, V., Muggia, L. et al. Marine Fungal Diversity and Dynamics in the Gulf of Trieste (Northern Adriatic Sea). Microb Ecol 87, 78 (2024). https://doi.org/10.1007/s00248-024-02394-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00248-024-02394-z