Abstract
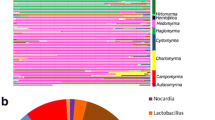
Microbes play a key role in the biology, ecology, and evolution of arthropods. Despite accumulating data on microbial communities in arthropods that feed on plants using piercing-sucking mouthparts, we still lack a comprehensive understanding of the composition and assembly factors of the microbiota, particularly in field-collected spider mites. Here, we applied 16S rRNA amplicon sequencing to investigate the characters of the bacterial community in 140 samples representing 420 mite individuals, belonging to eight Tetranychus species (Acari: Tetranychidae) collected from 26 sites in China. The results showed that the bacterial composition of spider mites varied significantly among different species, locations, and plants. The environment showed a significant influence on the bacterial community of spider mites, with different relative contributions. Latitude and precipitation were found to be the main factors influencing the bacterial community composition. The dissimilarity of bacterial community and geographical distance between mite locations were significantly correlated. The assembly of spider mite bacterial communities seemed to be mainly influenced by stochastic processes. Furthermore, the symbiont Cardinium was found to be important in shaping the microbiota of many Tetranychus species. The relative abundance of Cardinium was > 50% in T. viennensis, T. urticae G, T. urticae R, and T. turkestani. Removing Cardinium reads from our analysis significantly changed Shannon diversity index and weighted beta diversity in these species. Altogether, this study provides novel insights into bacterial diversity patterns that contribute to our knowledge of the symbiotic relationships between arthropods and their bacterial communities.






Similar content being viewed by others
Data Availability
The sequencing data have been deposited in the NCBI Sequence Read Archive (SRA) database with the BioProject number PRJNA555522. All supplementary files and associated dataset are available from Zenodo under the https://doi.org/10.5281/zenodo.8150083.
References
Stork NE (2018) How many species of insects and other terrestrial arthropods are there on earth? Annu Rev Entomol 63:31–45. https://doi.org/10.1146/annurev-ento-020117-043348
Wernegreen JJ (2012) Mutualism meltdown in insects: bacteria constrain thermal adaptation. Curr Opin Microbiol 15:255–262. https://doi.org/10.1016/j.mib.2012.02.001
Bing X, Gerlach J, Loeb G, Buchon N (2018) Nutrient-dependent impact of microbes on Drosophila suzukii development. mBio 9:e02199-e2117. https://doi.org/10.1128/mBio.02199-17
Lax S, Cardona C, Zhao D, Winton VJ, Goodney G, Gao P, Gottel N, Hartmann EM, Henry C, Thomas PM, Kelley ST, Stephens B, Gilbert JA (2019) Microbial and metabolic succession on common building materials under high humidity conditions. Nat Commun 10:1767. https://doi.org/10.1038/s41467-019-09764-z
Zhu Y-X, Song Z-R, Huo S-M, Yang K, Hong X-Y (2020) Variation in the microbiome of the spider mite Tetranychus truncatus with sex, instar and endosymbiont infection. FEMS Microbiol Ecol 96:fiaa004. https://doi.org/10.1093/femsec/fiaa004
Jones EW, Carlson JM, Sivak DA, Ludington WB (2022) Stochastic microbiome assembly depends on context. Proc Natl Acad Sci U S A 119:e2115877119. https://doi.org/10.1073/pnas.2115877119
Brinker P, Fontaine MC, Beukeboom LW, Falcao Salles J (2019) Host, symbionts, and the microbiome: the missing tripartite interaction. Trends Microbiol 27:480–488. https://doi.org/10.1016/j.tim.2019.02.002
Duan X-Z, Sun J-T, Wang L-T, Shu X-H, Guo Y, Keiichiro M, Zhu Y-X, Bing X-L, Hoffmann AA, Hong X-Y (2020) Recent infection by Wolbachia alters microbial communities in wild Laodelphax striatellus populations. Microbiome 8:104. https://doi.org/10.1186/s40168-020-00878-x
Fromont C, Adair KL, Douglas AE (2019) Correlation and causation between the microbiome, Wolbachia and host functional traits in natural populations of drosophilid flies. Mol Ecol 28:1826–1841. https://doi.org/10.1111/mec.15041
Dáder B, Then C, Berthelot E, Ducousso M, Ng JCK, Drucker M (2017) Insect transmission of plant viruses: multilayered interactions optimize viral propagation. Insect Sci 24:929–946. https://doi.org/10.1111/1744-7917.12470
Jing X, Wong ACN, Chaston JM, Colvin J, McKenzie CL, Douglas AE (2014) The bacterial communities in plant phloem-sap-feeding insects. Mol Ecol 23:1433–1444. https://doi.org/10.1111/mec.12637
Wang D, Huang Z, He H, Wei C (2018) Comparative analysis of microbial communities associated with bacteriomes, reproductive organs and eggs of the cicada Subpsaltria yangi. Arch Microbiol 200:227–235. https://doi.org/10.1007/s00203-017-1432-8
Walter DE, Proctor HC (2013) Mites: ecology, evolution & behaviour. Springer, Dordrecht
Bensoussan N, Santamaria ME, Zhurov V, Diaz I, Grbić M, Grbić V (2016) Plant-herbivore interaction: dissection of the cellular pattern of Tetranychus urticae feeding on the host plant. Front Plant Sci 7. https://doi.org/10.3389/fpls.2016.01105
Hong X-Y (2011) Agricultural Acarology. China Agriculture Press Co., Ltd, Beijing
Leeuwen TV, Dermauw W (2016) The molecular evolution of xenobiotic metabolism and resistance in chelicerate mites. Annu Rev Entomol 61:475–498. https://doi.org/10.1146/annurev-ento-010715-023907
Zélé F, Santos I, Olivieri I, Weill M, Duron O, Magalhães S (2018) Endosymbiont diversity and prevalence in herbivorous spider mite populations in South-Western Europe. FEMS Microbiol Ecol 94. https://doi.org/10.1093/femsec/fiy015
Zhu Y-X, Song Y-L, Zhang Y-K, Hoffmann AA, Zhou J-C, Sun J-T, Hong X-Y (2018) Incidence of facultative bacterial endosymbionts in spider mites associated with local environment and host plant. Appl Environ Microbiol 84:e02546-e2517. https://doi.org/10.1128/aem.02546-17
Zhang Y-K, Chen Y-T, Yang K, Qiao G-X, Hong X-Y (2016) Screening of spider mites (Acari: Tetranychidae) for reproductive endosymbionts reveals links between co-infection and evolutionary history. Sci Rep 6: 27900. https://doi.org/10.1038/srep27900https://www.nature.com/articles/srep27900#supplementary-information
Chaisiri K, McGarry JW, Morand S, Makepeace BL (2015) Symbiosis in an overlooked microcosm: a systematic review of the bacterial flora of mites. Parasitology 142:1152–1162. https://doi.org/10.1017/S0031182015000530
Zélé F, Weill M, Magalhães S (2018) Identification of spider-mite species and their endosymbionts using multiplex PCR. Exp Appl Acarol 74:123–138. https://doi.org/10.1007/s10493-018-0224-4
Xue W-X, Sun J-T, Witters J, Vandenhole M, Dermauw W, Bajda SA, Simma EA, Wybouw N, Villacis-Perez E, Van Leeuwen T (2023) Incomplete reproductive barriers and genomic differentiation impact the spread of resistance mutations between green- and red-colour morphs of a cosmopolitan mite pest. Mol Ecol 32:4278–4297. https://doi.org/10.1111/mec.16994
Villacis-Perez E, Snoeck S, Kurlovs AH, Clark RM, Breeuwer JAJ, Van Leeuwen T (2021) Adaptive divergence and post-zygotic barriers to gene flow between sympatric populations of a herbivorous mite. Commun Biol 4:853. https://doi.org/10.1038/s42003-021-02380-y
Suh E, Sim C, Park J-J, Cho K (2015) Inter-population variation for Wolbachia induced reproductive incompatibility in the haplodiploid mite Tetranychus urticae. Exp Appl Acarol 65:55–71. https://doi.org/10.1007/s10493-014-9846-3
Gotoh T, Sugasawa J, Noda H, Kitashima Y (2007) Wolbachia-induced cytoplasmic incompatibility in Japanese populations of Tetranychus urticae (Acari: Tetranychidae). Exp Appl Acarol 42:1–16. https://doi.org/10.1007/s10493-007-9072-3
Bing X-L, Lu Y-J, Xia C-B, Xia X, Hong X-Y (2020) Transcriptome of Tetranychus urticae embryos reveals insights into Wolbachia-induced cytoplasmic incompatibility. Insect Mol Biol 29:193–204. https://doi.org/10.1111/imb.12620
Wybouw N, Mortier F, Bonte D (2022) Interacting host modifier systems control Wolbachia-induced cytoplasmic incompatibility in a haplodiploid mite. Evolution Letters 6:255–265. https://doi.org/10.1002/evl3.282
Zhao D-X, Zhang X-F, Hong X-Y (2013) Host-symbiont interactions in spider mite Tetranychus truncatus doubly infected with Wolbachia and Cardinium. Environ Entomol 42:445–452. https://doi.org/10.1603/EN12354
Zhu L-Y, Zhang K-J, Zhang Y-K, Ge C, Gotoh T, Hong X-Y (2012) Wolbachia strengthens Cardinium-induced cytoplasmic incompatibility in the spider mite Tetranychus piercei McGregor. Curr Microbiol 65:516–523. https://doi.org/10.1007/s00284-012-0190-8
Zhu Y-X, Song Z-R, Song Y-L, Hong X-Y (2020) Double infection of Wolbachia and Spiroplasma alters induced plant defense and spider mite fecundity. Pest Manag Sci 76:3273–3281. https://doi.org/10.1002/ps.5886
Zhu Y-X, Song Z-R, Zhang Y-Y, Hoffmann AA, Hong X-Y (2021) Spider mites singly infected with either Wolbachia or Spiroplasma have reduced thermal tolerance. Front Microbiol 12. https://doi.org/10.3389/fmicb.2021.706321
Yang K, Chen H, Bing X-L, Xia X, Zhu Y-X, Hong X-Y (2021) Wolbachia and Spiroplasma could influence bacterial communities of the spider mite Tetranychus truncatus. Exp Appl Acarol 83:197–210. https://doi.org/10.1007/s10493-021-00589-4
Chen L, Sun J-T, Jin P-Y, Hoffmann AA, Bing X-L, Zhao D-S, Xue X-F, Hong X-Y (2020) Population genomic data in spider mites point to a role for local adaptation in shaping range shifts. Evol Appl 13:2821–2835. https://doi.org/10.1111/eva.13086
Bing X-L, Zhao D-S, Peng C-W, Huang H-J, Hong X-Y (2020) Similarities and spatial variations of bacterial and fungal communities in field rice planthopper (Hemiptera: Delphacidae) populations. Insect Sci 27:947–963. https://doi.org/10.1111/1744-7917.12782
Zhu Y-X, Song Z-R, Song Y-L, Zhao D-S, Hong X-Y (2020) The microbiota in spider mite feces potentially reflects intestinal bacterial communities in the host. Insect Sci 27:859–868. https://doi.org/10.1111/1744-7917.12716
Ge C, Ding X-L, Zhang J-P, Hong X-Y (2013) Tetranychus urticae (green form) on Gossypium hirsutum in China: two records confirmed by aedeagus morphology and RFLP analysis. Syst Appl Acarol 18:239–244. https://doi.org/10.11158/saa.18.3.6
Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M, Glöckner FO (2013) Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res 41:e1–e1. https://doi.org/10.1093/nar/gks808
Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, Bai Y, Bisanz JE, Bittinger K, Brejnrod A, Brislawn CJ, Brown CT, Callahan BJ, Caraballo-Rodríguez AM, Chase J, Cope EK, Da Silva R, Diener C, Dorrestein PC, Douglas GM, Durall DM, Duvallet C, Edwardson CF, Ernst M, Estaki M, Fouquier J, Gauglitz JM, Gibbons SM, Gibson DL, Gonzalez A, Gorlick K, Guo J, Hillmann B, Holmes S, Holste H, Huttenhower C, Huttley GA, Janssen S, Jarmusch AK, Jiang L, Kaehler BD, Kang KB, Keefe CR, Keim P, Kelley ST, Knights D, Koester I, Kosciolek T, Kreps J, Langille MGI, Lee J, Ley R, Liu Y-X, Loftfield E, Lozupone C, Maher M, Marotz C, Martin BD, McDonald D, McIver LJ, Melnik AV, Metcalf JL, Morgan SC, Morton JT, Naimey AT, Navas-Molina JA, Nothias LF, Orchanian SB, Pearson T, Peoples SL, Petras D, Preuss ML, Pruesse E, Rasmussen LB, Rivers A, Robeson MS, Rosenthal P, Segata N, Shaffer M, Shiffer A, Sinha R, Song SJ, Spear JR, Swafford AD, Thompson LR, Torres PJ, Trinh P, Tripathi A, Turnbaugh PJ, Ul-Hasan S, van der Hooft JJJ, Vargas F, Vázquez-Baeza Y, Vogtmann E, von Hippel M, Walters W, Wan Y, Wang M, Warren J, Weber KC, Williamson CHD, Willis AD, Xu ZZ, Zaneveld JR, Zhang Y, Zhu Q, Knight R, Caporaso JG (2019) Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 37:852–857. https://doi.org/10.1038/s41587-019-0209-9
Rognes T, Flouri T, Nichols B, Quince C, Mahé F (2016) VSEARCH: a versatile open source tool for metagenomics. PeerJ 4:e2584. https://doi.org/10.7717/peerj.2584
Amir A, McDonald D, Navas-Molina JA, Kopylova E, Morton JT, Xu ZZ, Kightley EP, Thompson LR, Hyde ER, Gonzalez A, Knight R (2017) Deblur rapidly resolves single-nucleotide community sequence patterns. mSystems 2: https://doi.org/10.1128/msystems.00191-00116. https://doi.org/10.1128/msystems.00191-16
Bokulich NA, Kaehler BD, Rideout JR, Dillon M, Bolyen E, Knight R, Huttley GA, Gregory Caporaso J (2018) Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 6:90. https://doi.org/10.1186/s40168-018-0470-z
Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO (2012) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596. https://doi.org/10.1093/nar/gks1219
Lozupone C, Lladser ME, Knights D, Stombaugh J, Knight R (2011) UniFrac: an effective distance metric for microbial community comparison. ISME J 5:169–172. https://doi.org/10.1038/ismej.2010.133
Oksanen J, Kindt R, Legendre P, O’Hara B, Stevens MHH, Oksanen MJ, Suggests M (2007) The vegan package. Community Ecol Package 10:631–637
McMurdie PJ, Holmes S (2013) phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. Plos One 8:e61217. https://doi.org/10.1371/journal.pone.0061217
R Team C (2022) R: a language and environment for statistical computing R Foundation for Statistical Computing. http://www.R-project.org/
Wickham H (2016) ggplot2: elegant graphics for data analysis. Springer
Sloan WT, Lunn M, Woodcock S, Head IM, Nee S, Curtis TP (2006) Quantifying the roles of immigration and chance in shaping prokaryote community structure. Environ Microbiol 8:732–740. https://doi.org/10.1111/j.1462-2920.2005.00956.x
Stegen JC, Lin X, Fredrickson JK, Chen X, Kennedy DW, Murray CJ, Rockhold ML, Konopka A (2013) Quantifying community assembly processes and identifying features that impose them. ISME J 7:2069–2079. https://doi.org/10.1038/ismej.2013.93
Kembel SW, Cowan PD, Helmus MR, Cornwell WK, Morlon H, Ackerly DD, Blomberg SP, Webb CO (2010) Picante: R tools for integrating phylogenies and ecology. Bioinformatics 26:1463–1464. https://doi.org/10.1093/bioinformatics/btq166
Auger P, Migeon A, Ueckermann EA, Tiedt L, Navajas M (2013) Evidence for synonymy between Tetranychus urticae and Tetranychus cinnabarinus (Acari, Prostigmata, Tetranychidae): Review and new data. Acarologia 53:383–415. https://doi.org/10.1051/acarologia/20132102
Jin P-Y, Tian L, Chen L (2012) Hong X-Y (2018) Spider mites of agricultural importance in China, with focus on species composition during the last decade (2008–2017). Syst Appl Acarol 23:2087–2098
Gawande SJ, Anandhan S, Ingle A, Roylawar P, Khandagale K, Gawai T, Jacobson A, Asokan R, Singh M (2019) Microbiome profiling of the onion thrips, Thrips tabaci Lindeman (Thysanoptera: Thripidae). Plos One 14:e0223281. https://doi.org/10.1371/journal.pone.0223281
Baumann P (2005) Biology of bacteriocyte-associated endosymbionts of plant sap-sucking insects. Annu Rev Microbiol 59:155–189. https://doi.org/10.1146/annurev.micro.59.030804.121041
Lemoine R, La Camera S, Atanassova R, Dédaldéchamp F, Allario T, Pourtau N, Bonnemain J-L, Laloi M, Coutos-Thévenot P, Maurousset L, Faucher M, Girousse C, Lemonnier P, Parrilla J, Durand M (2013) Source-to-sink transport of sugar and regulation by environmental factors. Front Plant Sci 4. https://doi.org/10.3389/fpls.2013.00272
Näpflin K, Schmid-Hempel P (2018) Host effects on microbiota community assembly. J Anim Ecol 87:331–340. https://doi.org/10.1111/1365-2656.12768
Van Opijnen T, Breeuwer J (1999) High temperatures eliminate Wolbachia, a cytoplasmic incompatibility inducing endosymbiont, from the two-spotted spider mite. Exp Appl Acarol 23:871–881
Sieber M, Pita L, Weiland-Bräuer N, Dirksen P, Wang J, Mortzfeld B, Franzenburg S, Schmitz RA, Baines JF, Fraune S, Hentschel U, Schulenburg H, Bosch TCG, Traulsen A (2019) Neutrality in the metaorganism. Plos Biol 17:e3000298. https://doi.org/10.1371/journal.pbio.3000298
Ge Y, Jing Z, Diao Q, He J-Z, Liu Y-J (2021) Host species and geography differentiate honeybee gut bacterial communities by changing the relative contribution of community assembly processes. mBio 12:e00751-00721. https://doi.org/10.1128/mBio.00751-21
Gottlieb Y, Ghanim M, Gueguen G, Kontsedalov S, Vavre F, Fleury F, Zchori-Fein E (2008) Inherited intracellular ecosystem: symbiotic bacteria share bacteriocytes in whiteflies. FASEB J 22:2591–2599. https://doi.org/10.1096/Fj.07-101162
Zchori-Fein E, Perlman SJ (2004) Distribution of the bacterial symbiont Cardinium in arthropods. Mol Ecol 13:2009–2016. https://doi.org/10.1046/j.1365-294X.2004.02203.x
Hubert J, Nesvorna M, Klimov PB, Erban T, Sopko B, Dowd SE, Scully ED, Bordenstein S (2021) Interactions of the intracellular bacterium Cardinium with its host, the house dust mite Dermatophagoides farinae, based on gene expression data. mSystems 6:e00916-00921. https://doi.org/10.1128/mSystems.00916-21
Zhang Y-K, Chen Y-T, Yang K, Hong X-Y (2016) A review of prevalence and phylogeny of the bacterial symbiont Cardinium in mites (subclass: Acari). Syst Appl Acarol 21:978–990. https://doi.org/10.11158/saa.21.7.11
Liu Y, Xie R-R, Hong X-Y (2010) Manipulation of symbiont Cardinium on the reproduction of the carmine spidermite, Tetranychus cinnabarinus (Boisduval) (Acari:Tetranychidae). Acta Entomol Sin 53:1233–1240
Gotoh T, Noda H, Ito S (2007) Cardinium symbionts cause cytoplasmic incompatibility in spider mites. Heredity 98:13–20. https://doi.org/10.1038/sj.hdy.6800881
Acknowledgements
We thank Prof. Jing-Tao Sun from Nanjing Agricultural University and Ary Hoffmann from University of Melbourne for the help on improving the manuscript. We thank the high-performance computing platform of Bioinformatics Center, Nanjing Agricultural University, for support with bioinformatical analysis.
Funding
This study was supported by the National Key R&D Program of China (No. 2022YFC2601000), National Natural Science Foundation of China (No. 32001905 and 32020103011), Natural Science Foundation of Jiangsu Province (No. BK20211213), and the Fundamental Research Funds for the Central Universities (No. KJQN202110). The funders played no role in study design, data collection, analysis and interpretation of data, or the writing of this manuscript.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection, and analysis were performed by Huan-Huan Liu, Lei Chen, Hui-Biao Shao, Shuo Gao, Xiao-Yue Hong, and Xiao-Li Bing. The first draft of the manuscript was written by Huan-Huan Liu and Xiao-Li Bing. All authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Supplementary Information
Below is the link to the electronic supplementary material.
248_2023_2314_MOESM2_ESM.csv
Supplementary file2 (CSV 36 KB) Sample information of spider mites and environmental factor information of collection sites
248_2023_2314_MOESM5_ESM.xlsx
Supplementary file5 (XLSX 12 KB) Mantel Test of the effect of Environmental factors on the bacterial Community of spider mites
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Liu, HH., Chen, L., Shao, HB. et al. Environmental Factors and the Symbiont Cardinium Influence the Bacterial Microbiome of Spider Mites Across the Landscape. Microb Ecol 87, 1 (2024). https://doi.org/10.1007/s00248-023-02314-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00248-023-02314-7