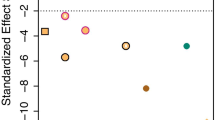
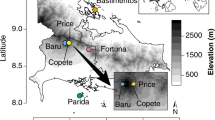
Abstract
While it is now well established that fungal community composition varies spatially at a variety of scales, temporal turnover of fungi is less well understood. Here we studied inter-annual community compositional changes of fungi in a rainforest tree canopy environment. We tracked fungal community shifts over 3 years in three substrate types (live bryophytes, dead bryophytes, and host tree bark) and compared these changes to amounts of community turnover seen at small spatial scales in the same system. The effect of substrate type on fungal community composition was stronger than that of sampling year, which was very small but significant. Although levels of temporal turnover varied among substrates, with greater turnover in live bryophytes than other substrates, the amount of turnover from year to year was comparable to what is seen at spatial distances between 5 and 9 cm for the same substrate. Stability of communities was largely driven by a few fungi with high relative abundances. A majority of fungal occurrences were at low relative abundances (≤ 0.1%). These fungi tended to be short lived and persisted to following years ≤ 50% of the time, depending on substrate. Their presence and persistence are likely impacted by stochastic processes like dispersal limitation and disturbance. Most samples contained only one or a few fungi at high relative abundance (≥ 10%) that persisted half or more of the time. These more abundant and persistent fungi are expected to have sustained functional interactions within the canopy ecosystem.






Similar content being viewed by others
Data Availability
The DNA sequence data generated and analyzed during this study are available in NCBI SRA accession PRJNA762332. Scripts used for data analysis and figure construction can be accessed at https://github.com/k-cook-fun/Tapanti-fungi-temporal
References
Bahram M, Kõljalg U, Courty P-E et al (2013) The distance decay of similarity in communities of ectomycorrhizal fungi in different ecosystems and scales. J Ecol 101:1335–1344. https://doi.org/10.1111/1365-2745.12120
Oono R, Rasmussen A, Lefèvre E (2017) Distance decay relationships in foliar fungal endophytes are driven by rare taxa: distance decay in fungal endophytes. Environ Microbiol 19:2794–2805. https://doi.org/10.1111/1462-2920.13799
Bahram M, Peay KG, Tedersoo L (2015) Local-scale biogeography and spatiotemporal variability in communities of mycorrhizal fungi. New Phytol 205:1454–1463. https://doi.org/10.1111/nph.13206
Chaudhary VB, O’Dell TE, Rillig MC, Johnson NC (2014) Multiscale patterns of arbuscular mycorrhizal fungal abundance and diversity in semiarid shrublands. Fungal Ecol 12:32–43. https://doi.org/10.1016/j.funeco.2014.06.003
Anderson JB, Bruhn JN, Kasimer D et al (2018) Clonal evolution and genome stability in a 2,500-year-old fungal individual. Proc R Soc B 285:20182233. https://doi.org/10.1098/rspb.2018.2233
Godbold DL, Hoosbeek MR, Lukac M et al (2006) Mycorrhizal hyphal turnover as a dominant process for carbon input into soil organic matter. Plant Soil 281:15–24. https://doi.org/10.1007/s11104-005-3701-6
Treseder KK, Allen MF, Ruess RW et al (2005) Lifespans of fungal rhizomorphs under nitrogen fertilization in a pinyon-juniper woodland. Plant Soil 270:249–255. https://doi.org/10.1007/s11104-004-1559-7
Staddon PL, Ramsey CB, Ostle N et al (2003) Rapid turnover of hyphae of mycorrhizal fungi determined by AMS microanalysis of 14C. Science 300:1138–1140. https://doi.org/10.1126/science.1084269
Pepe A, Giovannetti M, Sbrana C (2018) Lifespan and functionality of mycorrhizal fungal mycelium are uncoupled from host plant lifespan. Sci Rep 8:10235. https://doi.org/10.1038/s41598-018-28354-5
Peay KG, Bruns TD (2014) Spore dispersal of basidiomycete fungi at the landscape scale is driven by stochastic and deterministic processes and generates variability in plant-fungal interactions. New Phytol 204:180–191. https://doi.org/10.1111/nph.12906
Kivlin SN, Hawkes CV (2020) Spatial and temporal turnover of soil microbial communities is not linked to function in a primary tropical forest. Ecology 101:e02985. https://doi.org/10.1002/ecy.2985
Kyaschenko J, Clemmensen KE, Hagenbo A et al (2017) Shift in fungal communities and associated enzyme activities along an age gradient of managed Pinus sylvestris stands. ISME J 11:863–874. https://doi.org/10.1038/ismej.2016.184
Barrett LG, Kniskern JM, Bodenhausen N et al (2009) Continua of specificity and virulence in plant host–pathogen interactions: causes and consequences. New Phytol 183:513–529. https://doi.org/10.1111/j.1469-8137.2009.02927.x
Shefferson RP, WEIß M, Kull T, Taylor DL (2005) High specificity generally characterizes mycorrhizal association in rare lady’s slipper orchids, genus Cypripedium. Mol Ecol 14:613–626.https://doi.org/10.1111/j.1365-294X.2005.02424.x
Richard F, Millot S, Gardes M, Selosse M-A (2005) Diversity and specificity of ectomycorrhizal fungi retrieved from an old-growth Mediterranean forest dominated by Quercus ilex. New Phytol 166:1011–1023. https://doi.org/10.1111/j.1469-8137.2005.01382.x
Ishida TA, Nara K, Hogetsu T (2007) Host effects on ectomycorrhizal fungal communities: insight from eight host species in mixed conifer-broadleaf forests. New Phytol 174:430–440. https://doi.org/10.1111/j.1469-8137.2007.02016.x
Opik M, Metsis M, Daniell TJ et al (2009) Large-scale parallel 454 sequencing reveals host ecological group specificity of arbuscular mycorrhizal fungi in a boreonemoral forest. New Phytol 184:424–437. https://doi.org/10.1111/j.1469-8137.2009.02920.x
Osiewacz HD (2002) Genes, mitochondria and aging in filamentous fungi. Ageing Res Rev 1:425–442. https://doi.org/10.1016/S1568-1637(02)00010-7
Ovaskainen O, Schigel D, Ali-Kovero H et al (2013) Combining high-throughput sequencing with fruit body surveys reveals contrasting life-history strategies in fungi. ISME J 7:1696–1709. https://doi.org/10.1038/ismej.2013.61
Maynard DS, Bradford MA, Covey KR et al (2019) Consistent trade-offs in fungal trait expression across broad spatial scales. Nat Microbiol 4:846–853. https://doi.org/10.1038/s41564-019-0361-5
Lilleskov EA, Bruns TD (2003) Root colonization dynamics of two ectomycorrhizal fungi of contrasting life history strategies are mediated by addition of organic nutrient patches. New Phytol 159:141–151. https://doi.org/10.1046/j.1469-8137.2003.00794.x
IJdo M, Schtickzelle N, Cranenbrouck S, Declerck S, (2010) Do arbuscular mycorrhizal fungi with contrasting life-history strategies differ in their responses to repeated defoliation? FEMS Microbiol Ecol 72:114–122. https://doi.org/10.1111/j.1574-6941.2009.00829.x
López-García Á, Palenzuela J, Barea JM, Azcón-Aguilar C (2014) Life-history strategies of arbuscular mycorrhizal fungi determine succession into roots of Rosmarinus officinalis L., a characteristic woody perennial plant species from Mediterranean ecosystems. Plant Soil 379:247–260. https://doi.org/10.1007/s11104-014-2060-6
Hiscox J, O’Leary J, Boddy L (2018) Fungus wars: basidiomycete battles in wood decay. Stud Mycol 89:117–124. https://doi.org/10.1016/j.simyco.2018.02.003
Bainard LD, Bainard JD, Hamel C, Gan Y (2014) Spatial and temporal structuring of arbuscular mycorrhizal communities is differentially influenced by abiotic factors and host crop in a semi-arid prairie agroecosystem. FEMS Microbiol Ecol 88:333–344. https://doi.org/10.1111/1574-6941.12300
Taylor DL, Herriott IC, Stone KE et al (2010) Structure and resilience of fungal communities in Alaskan boreal forest soilsThis article is one of a selection of papers from The Dynamics of Change in Alaska’s Boreal Forests: Resilience and Vulnerability in Response to Climate Warming. Can J For Res 40:1288–1301. https://doi.org/10.1139/X10-081
Montero Sommerfeld H, Díaz LM, Alvarez M et al (2013) High winter diversity of arbuscular mycorrhizal fungal communities in shallow and deep grassland soils. Soil Biol Biochem 65:236–244. https://doi.org/10.1016/j.soilbio.2013.06.002
Voříšková J, Brabcová V, Cajthaml T, Baldrian P (2014) Seasonal dynamics of fungal communities in a temperate oak forest soil. New Phytol 201:269–278. https://doi.org/10.1111/nph.12481
Kivlin SN, Hawkes CV (2016) Tree species, spatial heterogeneity, and seasonality drive soil fungal abundance, richness, and composition in Neotropical rainforests. Environ Microbiol 18:4662–4673. https://doi.org/10.1111/1462-2920.13342
Reyes HA, Ferreira PFA, Silva LC et al (2019) Arbuscular mycorrhizal fungi along secondary forest succession at the eastern periphery of Amazonia: seasonal variability and impacts of soil fertility. Appl Soil Ecol 136:1–10. https://doi.org/10.1016/j.apsoil.2018.12.013
Averill C, Cates LL, Dietze MC, Bhatnagar JM (2019) Spatial vs. temporal controls over soil fungal community similarity at continental and global scales. ISME J 13:2082–2093. https://doi.org/10.1038/s41396-019-0420-1
Izzo A, Agbowo J, Bruns TD (2005) Detection of plot-level changes in ectomycorrhizal communities across years in an old-growth mixed-conifer forest. New Phytol 166:619–630. https://doi.org/10.1111/j.1469-8137.2005.01354.x
Cook K, Sharma J, Taylor AD et al (2022) Epiphytic fungal communities vary by substrate type and at submetre spatial scales. Mol Ecol 31:1879–1891. https://doi.org/10.1111/mec.16358
Taylor DL, Walters WA, Lennon NJ et al (2016) Accurate estimation of fungal diversity and abundance through improved lineage-specific primers optimized for Illumina amplicon sequencing. Appl Environ Microbiol 82:7217–7226. https://doi.org/10.1128/AEM.02576-16
Edgar RC (2013) UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10:996–998. https://doi.org/10.1038/nmeth.2604
Martin M (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17:10–12. https://doi.org/10.14806/ej.17.1.200
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267. https://doi.org/10.1128/AEM.00062-07
Nilsson RH, Larsson K-H, Taylor AFS et al (2019) The UNITE database for molecular identification of fungi: handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res 47:D259–D264. https://doi.org/10.1093/nar/gky1022
R Core Team (2018) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/
Oksanen J, Blanchet FG, Friendly M et al (2019) vegan: Community Ecology Package. R package version 2.5-4. https://CRAN.R-project.org/package=vegan
Blanchet FG, Legendre P, Borcard D (2008) Forward selection of explanatory variables. Ecology 89:2623–2632. https://doi.org/10.1890/07-0986.1
McMurdie PJ, Holmes S (2013) phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 8:e61217
Bates D, Mächler M, Bolker B, Walker S (2015) Fitting linear mixed-effects models using lme4. J Stat Softw 67:1–48. https://doi.org/10.18637/jss.v067.i01
Lenth R (2019) emmeans: estimated marginal means, aka least-squares means. R package version 1.3.3. https://CRAN.R-project.org/package=emmeans
Baselga A (2010) Partitioning the turnover and nestedness components of beta diversity: partitioning beta diversity. Glob Ecol Biogeogr 19:134–143. https://doi.org/10.1111/j.1466-8238.2009.00490.x
Baselga A, Orme D, Villeger S et al (2018) betapart: partitioning beta diversity into turnover and nestedness components. R package version 1.5.1. https://CRAN.R-project.org/package=betapart
Banerjee RD, Sen SP (1979) Antibiotic activity of bryophytes. Bryologist 82:141–153. https://doi.org/10.2307/3242073
Sabovljević A, Soković M, Glamočlija J et al (2011) Bio-activities of extracts from some axenically farmed and naturally grown bryophytes. JMPR 5:565–571. https://doi.org/10.5897/JMPR.9000290
Fukami T, Dickie IA, Paula Wilkie J et al (2010) Assembly history dictates ecosystem functioning: evidence from wood decomposer communities: carbon dynamics and fungal community assembly. Ecol Lett 13:675–684. https://doi.org/10.1111/j.1461-0248.2010.01465.x
Kennedy PG, Peay KG, Bruns TD (2009) Root tip competition among ectomycorrhizal fungi: are priority effects a rule or an exception? Ecology 90:2098–2107. https://doi.org/10.1890/08-1291.1
Chapela IH, Boddy L (1988) The fate of early fungal colonizers in beech branches decomposing on the forest floor. FEMS Microbiol Ecol 4:273–283. https://doi.org/10.1111/j.1574-6968.1988.tb02673.x-i1
Brislawn CJ, Graham EB, Dana K et al (2019) Forfeiting the priority effect: turnover defines biofilm community succession. ISME J 13:1865–1877. https://doi.org/10.1038/s41396-019-0396-x
Funding
This material is based upon work supported by the National Science Foundation under Grant No. DEB-1355155. High-performance computing resources used in the work was provided by the UNM Center for Advanced Research Computing, supported in part by the National Science Foundation.
Author information
Authors and Affiliations
Contributions
All authors contributed to study conception and design and sample collection. The first draft of the manuscript was written by Kel Cook. All authors reviewed and edited the manuscript. All authors read and approved of the final manuscript.
Corresponding author
Ethics declarations
Conflict of Interests
The authors declare no competing interests.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Cook, K., Taylor, A.D., Sharma, J. et al. Inter-annual Persistence of Canopy Fungi Driven by Abundance Despite High Spatial Turnover. Microb Ecol 86, 261–270 (2023). https://doi.org/10.1007/s00248-022-02104-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-022-02104-7