Abstract
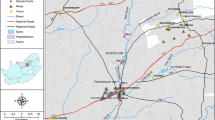
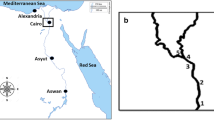
Environmental changes and human activities can alter the structure and diversity of aquatic microbial communities. In this work, we analyzed the bacterial community dynamics of an urban stream to understand how these factors affect the composition of river microbial communities. Samples were taken from a stream situated in Buenos Aires, Argentina, which flows through residential, peri-urban horticultural, and industrial areas. For sampling, two stations were selected: one influenced by a series of industrial waste treatment plants and horticultural farms (PL), and the other influenced by residential areas (R). Microbial communities were analyzed by sequence analysis of 16S rRNA gene amplicons along an annual cycle. PL samples showed high nutrient content compared with R samples. The diversity and richness of the R site were more affected by seasonality than those of the PL site. At the amplicon sequence variants level, beta diversity analysis showed a differentiation between cool-season (fall and winter) and warm-season (spring and summer) samples, as well as between PL and R sites. This demonstrated that there is spatial and temporal heterogeneity in the composition of the bacterial community, which should be considered if a bioremediation strategy is applied. The taxonomic composition analysis also revealed a differential seasonal cycle of phototrophs and chemoheterotrophs between the sampling sites, as well as different taxa associated with each sampling site. This analysis, combined with a comparative analysis of global rivers, allowed us to determine the genera Arcobacter, Simplicispira, Vogesella, and Sphingomonas as potential bioindicators of anthropogenic disturbance.






Similar content being viewed by others
References
Martí E, Riera JL, Sabater F (2009) Effects of wastewater treatment plants on stream nutrient dynamics under water scarcity conditions. Handbook Environ Chem 173–195
Drury B, Rosi-Marshall E, Kelly JJ (2013) Wastewater treatment effluent reduces the abundance and diversity of benthic bacterial communities in urban and suburban rivers. Appl Environ Microbiol 79:1897–1905
Köchling T, Sanz JL, Galdino L et al (2017) Impact of pollution on the microbial diversity of a tropical river in an urbanized region of northeastern Brazil. Int Microbiol 20:11–24. https://doi.org/10.2436/20.1501.01.281
Fisher JC, Newton RJ, Dila DK, McLellan SL (2015) Urban microbial ecology of a freshwater estuary of Lake Michigan. Elementa Sci Anthropocene 3:000064
Talabi AO, Kayode TJ (2019) Groundwater pollution and remediation. J Water Resour Prot 11:1–19
Vandermaesen J, Horemans B, Bers K et al (2016) Application of biodegradation in mitigating and remediating pesticide contamination of freshwater resources: state of the art and challenges for optimization. Appl Microbiol Biotechnol 100:7361–7376. https://doi.org/10.1007/s00253-016-7709-z
Vujnovič R (1990) Urban development in the Danubian Basin and its effects on water quality–aspects and trends. Water Sci Technol 22:281–286. https://doi.org/10.2166/wst.1990.0041
Singh M, Ansari AA, Müller G, Singh IB (1997) Heavy metals in freshly deposited sediments of the Gomati River (a tributary of the Ganga River): effects of human activities. Environ Geol 29:246–252
Ternus RZ, de Souza-Franco GM, Anselmini MEK et al (2011) Influence of urbanisation on water quality in the basin of the upper Uruguay River in western Santa Catarina, Brazil. Acta Limnol Bras 23:189–199
Paudyal R, Kang S, Sharma CM, et al (2016) Variations of the physicochemical parameters and metal levels and their risk assessment in urbanized Bagmati River, Kathmandu, Nepal. J Chem Chem Eng 2016. https://doi.org/10.1155/2016/6025905
Vatanpour N, Malvandi AM, Talouki HH et al (2020) Impact of rapid urbanization on the surface water’s quality: a long-term environmental and physicochemical investigation of Tajan river, Iran (2007–2017). Environ Sci Pollut Res 27:8439–8450
Wakelin SA, Colloff MJ, Kookana RS (2008) Effect of wastewater treatment plant effluent on microbial function and community structure in the sediment of a freshwater stream with variable seasonal flow. Appl Environ Microbiol 74:2659–2668. https://doi.org/10.1128/AEM.02348-07
Zhang X, Gu Q, Long X-E et al (2016) Anthropogenic activities drive the microbial community and its function in urban river sediment. J Soils Sediments 16:716–725
Christiansen NA, Fryirs KA, Green TJ, Hose GC (2019) The impact of urbanisation on community structure, gene abundance and transcription rates of microbes in upland swamps of Eastern Australia. PLoS ONE 14:e0213275. https://doi.org/10.1371/journal.pone.0213275
Beattie RE, Bandla A, Swarup S, Hristova KR (2020) Freshwater sediment microbial communities are not resilient to disturbance from agricultural land runoff. Front Microbiol 11:539921. https://doi.org/10.3389/fmicb.2020.539921
Li K, Hu J, Li T, et al (2021) Microbial abundance and diversity investigations along rivers: current knowledge and future directions. WIREs Water 8. https://doi.org/10.1002/wat2.1547
Ronco A, Peluso L, Jurado M, et al (2008) Screening of sediment pollution in tributaries from the southwestern coast of the Río de la Plata estuary. Latin American journal of sedimentology and basin analysis 15:
Salibián A (2006) Ecotoxicological Assessment of the highly polluted reconquista river of Argentina. In: Ware GW, Nigg HN, Doerge DR (eds) Reviews of environmental contamination and toxicology. Springer, New York, New York, NY, pp 35–65
López OCF, Duverne LB, Mazieres JO, Salibián A (2013) Microbiological pollution of surface water in the upper-middle basin of the Reconquista river (Argentina): 2010–2011 monitoring. Int J Environ Health 6:276
Nader GM, Proaño PVS, Cicerone DS (2013) Water quality assessment of a polluted urban river. Int J Environ Health 6:307–319. https://doi.org/10.1504/IJENVH.2013.056972
Ossana NA, Castañé PM, Salibián A (2013) Use of Lithobates catesbeianus tadpoles in a multiple biomarker approach for the assessment of water quality of the Reconquista River (Argentina). Arch Environ Contam Toxicol 65:486–497. https://doi.org/10.1007/s00244-013-9920-6
Baudou FG, Ossana NA, Castañé PM et al (2019) Use of integrated biomarker indexes for assessing the impact of receiving waters on a native neotropical teleost fish. Sci Total Environ 650:1779–1786. https://doi.org/10.1016/j.scitotenv.2018.09.342
Castilla V, Canevaro S, López MB (2021) Migración, degradación ambiental y percepciones del riesgo en la cuenca del río Reconquista (Buenos Aires, Argentina). Rev.estud.soc 41–57. https://doi.org/10.7440/res76.2021.04
Vullo DL, Ceretti HM, Hughes EA et al (2005) Indigenous heavy metal multiresistant microbiota of Las Catonas stream. Environ Monit Assess 105:81–97. https://doi.org/10.1007/s10661-005-3157-4
Cantera CG, Scasso RA, Tufo A, et al (2018) Mobility of trace elements between the river water, the sediments, and the pore water of Las Catonas Stream, Buenos Aires Province, Argentina. Environ Earth Sci 77
RaigerIustman LJ, Almasqué FJ, Vullo DL (2021) Microbiota diversity change as quality indicator of soils exposed to intensive periurban agriculture. Curr Microbiol 78:338–346. https://doi.org/10.1007/s00284-020-02298-4
Ibarbalz FM, Pérez MV, Figuerola ELM, Erijman L (2014) The bias associated with amplicon sequencing does not affect the quantitative assessment of bacterial community dynamics. PLoS ONE 9:e99722
Callahan BJ, McMurdie PJ, Rosen MJ et al (2016) DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583. https://doi.org/10.1038/nmeth.3869
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73. https://doi.org/10.1128/AEM.00062-07
Oksanen J, Blanchet FG, Kindt R, et al (2013) Community ecology package. R package version 2:
Roberts DW, Roberts MDW (2016) Package “labdsv.” Ordination and Multivariate 775:
Conway JR, Lex A, Gehlenborg N (2017) UpSetR: an R package for the visualization of intersecting sets and their properties. Bioinformatics 33:2938–2940. https://doi.org/10.1093/bioinformatics/btx364
Mitchell AL, Almeida A, Beracochea M et al (2020) MGnify: the microbiome analysis resource in 2020. Nucleic Acids Res 48:D570–D578. https://doi.org/10.1093/nar/gkz1035
Meyer F, Paarmann D, D’Souza M et al (2008) The metagenomics RAST server - a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics 9:386. https://doi.org/10.1186/1471-2105-9-386
Xie H, Ringler C (2017) Agricultural nutrient loadings to the freshwater environment: the role of climate change and socioeconomic change. Environ Res Lett 12:104008. https://doi.org/10.1088/1748-9326/aa8148
Marti E, Aumatell J, Godé L et al (2004) Nutrient retention efficiency in streams receiving inputs from wastewater treatment plants. J Environ Qual 33:285–293. https://doi.org/10.2134/jeq2004.2850
Uysal Z, Köksalan İ (2006) The annual cycle of Synechococcus (cyanobacteria) in the northern Levantine Basin shelf waters (Eastern Mediterranean). Mar Ecol 27:187–197. https://doi.org/10.1111/j.1439-0485.2006.00105.x
Ferrera I, Borrego CM, Salazar G, Gasol JM (2014) Marked seasonality of aerobic anoxygenic phototrophic bacteria in the coastal NW Mediterranean Sea as revealed by cell abundance, pigment concentration and pyrosequencing of pufM gene. Environ Microbiol 16:2953–2965. https://doi.org/10.1111/1462-2920.12278
Roeselers G, van Loosdrecht MCM, Muyzer G (2008) Phototrophic biofilms and their potential applications. J Appl Phycol 20:227–235. https://doi.org/10.1007/s10811-007-9223-2
Li M, Jain S, Dick GJ (2016) Genomic and transcriptomic resolution of organic matter utilization among deep-sea bacteria in guaymas basin hydrothermal plumes. Front Microbiol 7:1125. https://doi.org/10.3389/fmicb.2016.01125
Anandan R, Dharumadurai D, Manogaran GP (2016) An introduction to actinobacteria. In: Dhanasekaran D, Jiang Y (eds) Actinobacteria. IntechOpen, Rijeka
Zhao D, Cao X, Huang R et al (2017) Variation of bacterial communities in water and sediments during the decomposition of Microcystis biomass. PLoS ONE 12:e0176397. https://doi.org/10.1371/journal.pone.0176397
Harke MJ, Steffen MM, Gobler CJ et al (2016) A review of the global ecology, genomics, and biogeography of the toxic cyanobacterium, Microcystis spp. Harmful Algae 54:4–20. https://doi.org/10.1016/j.hal.2015.12.007
U. s. Environmental Protection Agency (2015) Literature review of contaminants in livestock and poultry manure and implications for water quality. CreateSpace
Tian L, Wang L (2020) A meta-analysis of microbial community structures and associated metabolic potential of municipal wastewater treatment plants in global scope. Environ Pollut 263:114598. https://doi.org/10.1016/j.envpol.2020.114598
Lee KCY, Dunfield PF, Stott MB (2014) The phylum Armatimonadetes. The prokaryotes. Springer, Berlin, Heidelberg, pp 447–458
Collado L, Figueras MJ (2011) Taxonomy, epidemiology, and clinical relevance of the genus Arcobacter. Clin Microbiol Rev 24:174–192. https://doi.org/10.1128/CMR.00034-10
Kristensen JM, Nierychlo M, Albertsen M, Nielsen PH (2020) Bacteria from the genus are abundant in effluent from wastewater treatment plants. Appl Environ Microbiol 86. https://doi.org/10.1128/AEM.03044-19
Lu S, Ryu SH, Chung BS et al (2007) Simplicispira limi sp. nov., isolated from activated sludge. Int J Syst Evol Microbiol 57:31–34. https://doi.org/10.1099/ijs.0.64566-0
Dawson KS, Schaperdoth I, Freeman KH, Macalady JL (2013) Anaerobic biodegradation of the isoprenoid biomarkers pristane and phytane. Org Geochem 65:118–126. https://doi.org/10.1016/j.orggeochem.2013.10.010
Numberger D, Ganzert L, Zoccarato L et al (2019) Characterization of bacterial communities in wastewater with enhanced taxonomic resolution by full-length 16S rRNA sequencing. Sci Rep 9:9673. https://doi.org/10.1038/s41598-019-46015-z
Krieg NR (2015) Vogesella. Bergey’s Manual of Systematics of Archaea and Bacteria 1–3
Salcher MM, Neuenschwander SM, Posch T, Pernthaler J (2015) The ecology of pelagic freshwater methylotrophs assessed by a high-resolution monitoring and isolation campaign. ISME J 9:2442–2453. https://doi.org/10.1038/ismej.2015.55
Balmonte JP, Arnosti C, Underwood S et al (2016) Riverine bacterial communities reveal environmental disturbance signatures within the Betaproteobacteria and Verrucomicrobia. Front Microbiol 7:1441. https://doi.org/10.3389/fmicb.2016.01441
Acknowledgements
DFDP, AGT, NIL, RMH, and LJRI are CONICET researchers, while FAV, AMP, and DBR are CONICET doctoral fellows. Sampling permissions were obtained from IMDEL (Instituto Municipal de Desarrollo Económico Local) and the Municipality of Moreno. We especially thank Mrs. Verónica Eugenia Zimmer from IMDEL for her collaboration during the field campaigns.
Funding
This work was supported by the Consejo Nacional de Investigaciones Científicas y Tecnológicas (CONICET), grant PUE2016 22920160100064CO (LJRI) and the University of Buenos Aires: grants 20020170200338BA (RMH) and 20020170100310BA (NIL).
Author information
Authors and Affiliations
Contributions
Conceptualization: Federico A. Vignale, Nancy I. López, Laura J. Raiger Iustman; data curation: Federico A. Vignale, Agustín M. Pardo; formal analysis: Federico A. Vignale, Agustín M. Pardo, Daissy Bernal Rey, Facundo J. Almasqué; funding acquisition: Renata Menéndez Helman, Nancy I. López, Laura J. Raiger Iustman; investigation: Daissy Bernal Rey, Facundo J. Almasqué, José G. Ibarra, Renata Menéndez Helman, Laura J. Raiger Iustman; project administration: Laura J. Raiger Iustman; resources: Renata Menéndez Helman, Nancy I. López, Darío Fernández Do Porto, Adrián G. Turjanski, Laura J. Raiger Iustman; software: Agustín M. Pardo; supervision: Laura J. Raiger Iustman; validation: Federico A. Vignale, Nancy I. López, Laura J. Raiger Iustman; visualization: Federico A. Vignale, Laura J. Raiger Iustman; writing: Federico A. Vignale (original draft, reviewing, and editing), Daissy Bernal Rey (original draft, reviewing, and editing), Agustín M. Pardo, Darío Fernández Do Porto (review), Adrián G. Turjanski (review), Nancy I. López (original draft, reviewing, and editing), Renata Menéndez Helman (original draft, reviewing, and editing), Laura J. Raiger Iustman (original draft, reviewing, and editing).
Corresponding author
Ethics declarations
Consent to Participation and Publication
All authors and their institution consent their participation in the present work and the final manuscript publication.
Conflict of Interest
The authors declare no competing interests.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Vignale, F.A., Bernal Rey, D., Pardo, A.M. et al. Spatial and Seasonal Variations in the Bacterial Community of an Anthropogenic Impacted Urban Stream. Microb Ecol 85, 862–874 (2023). https://doi.org/10.1007/s00248-022-02055-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-022-02055-z