Abstract
Insect gut bacteria have a significant impact on host biology, which has a favorable or negative impact on insect fitness. The walnut leaf beetle (Gastrolina depressa) is a notorious pest in China, causing severe damage to Juglandaceae trees including Juglans regia and Pterocarya rhoifolia. To date, however, we know surprisingly little about the gut microbiota of G. depressa. This study used a high-throughput sequencing platform to investigate the gut bacterial community of G. depressa throughout its life cycle, including the 1st, 2nd, and 3rd instar larvae, as well as male, female, and pre-pregnant female adults. Our results showed that the diversity of the gut bacterial community in larvae was generally higher than that in adults, and young larvae (1st and 2nd larvae) possessed the most diversified and abundant community. Principal coordinate analysis results showed that the gut microbiota of adults cluster together, which is independent of the 1st and 2nd instar larvae. The main phyla were Proteobacteria and Firmicutes in the microbial community of G. depressa, while the dominant genera were Enterobacter, Rosenbergiella, Erwinia, Pseudomonas, and Lactococcus. The gut bacteria of G. depressa were mostly enriched in metabolic pathways (carbohydrate metabolism and amino acid metabolism) as revealed by functional prediction. This study contributes to a better knowledge of G. depressa’s gut microbiota and its potential interactions with the host insect, facilitating the development of a microbial-based pest management strategy.





Similar content being viewed by others
Data Availability
The raw full-length 16S rRNA gene sequences generated in the present study were deposited in the Sequence Read Archive (SRA) database (https:// submit.ncbi.nlm.nih.gov/subs/sra/) under accession number PRJNA768929.
References
Moran NA, Ochman H, Hammer TJ (2019) Evolutionary and ecological consequences of gut microbial communities. Annu Rev Ecol Evol Syst 50(1):451–475. https://doi.org/10.1146/annurev-ecolsys-110617-062453
Engel P, Moran NA (2013) The gut microbiota of insects - diversity in structure and function. FEMS Microbiol Rev 37(5):699–735. https://doi.org/10.1111/1574-6976.12025
Consuegra J, Grenier T, Akherraz H, Rahioui I, Gervais H, da Silva P, Leulier F (2020) Metabolic cooperation among commensal bacteria supports Drosophila Juvenile growth under nutritional stress. iScience 23(6):101232. https://doi.org/10.1016/j.isci.2020.101232
Tokuda G, Mikaelyan A, Fukui C, Matsuura Y, Watanabe H, Fujishima M, Brune A (2018) Fiber-associated spirochetes are major agents of hemicellulose degradation in the hindgut of wood-feeding higher termites. Proc Natl Acad Sci U S A 115(51):E11996–E12004. https://doi.org/10.1073/pnas.1810550115
Zheng H, Powell JE, Steele MI, Dietrich C, Moran NA (2017) Honeybee gut microbiota promotes host weight gain via bacterial metabolism and hormonal signaling. Proc Natl Acad Sci U S A 114(18):4775–4780. https://doi.org/10.1073/pnas.1701819114
Storelli G, Defaye A, Erkosar B, Hols P, Royet J, Leulier F (2011) Lactobacillus plantarum promotes Drosophila systemic growth by modulating hormonal signals through TOR-dependent nutrient sensing. Cell Metab 14(3):403–414. https://doi.org/10.1016/j.cmet.2011.07.012
Berasategui A, Salem H, Paetz C, Santoro M, Gershenzon J, Kaltenpoth M, Schmidt A (2017) Gut microbiota of the pine weevil degrades conifer diterpenes and increases insect fitness. Mol Ecol 26(15):4099–4110. https://doi.org/10.1111/mec.14186
Zhou F, Wu X, Xu L, Guo S, Chen G, Zhang X (2019) Repressed Beauveria bassiana infections in Delia antiqua due to associated microbiota. Pest Manag Sci 75(1):170–179. https://doi.org/10.1002/ps.5084
Zhou F, Xu L, Wu X, Zhao X, Liu M, Zhang X (2020) Symbiotic Bacterium-derived organic acids protect larvae from entomopathogenic fungal infection. mSystems 5(6). https://doi.org/10.1128/mSystems.00778-20
Sharon G, Segal D, Ringo JM, Hefetz A, Zilber-Rosenberg I, Rosenberg E (2010) Commensal bacteria play a role in mating preference of Drosophila melanogaster. Proc Natl Acad Sci USA 107(46):20051–20056. https://doi.org/10.1073/pnas.1009906107
Schretter CE, Vielmetter J, Bartos I, Marka Z, Marka S, Argade S, Mazmanian SK (2018) A gut microbial factor modulates locomotor behaviour in Drosophila. Nature 563(7731):402–406. https://doi.org/10.1038/s41586-018-0634-9
Jia Y, Jin S, Hu K, Geng L, Han C, Kang R, Pang Y, Ling E, Tan EK, Pan Y, Liu W (2021) Gut microbiome modulates Drosophila aggression through octopamine signaling. Nat Commun 12(1):2698. https://doi.org/10.1038/s41467-021-23041-y
Xu L, Xu S, Sun L, Zhang Y, Luo J, Bock R, Zhang J (2021) Synergistic action of the gut microbiota in environmental RNA interference in a leaf beetle. Microbiome 9(1):98. https://doi.org/10.1186/s40168-021-01066-1
Wei G, Lai Y, Wang G, Chen H, Li F, Wang S (2017) Insect pathogenic fungus interacts with the gut microbiota to accelerate mosquito mortality. Proc Natl Acad Sci U S A 114(23):5994–5999. https://doi.org/10.1073/pnas.1703546114
Abraham NM, Liu L, Jutras BL, Yadav AK, Narasimhan S, Gopalakrishnan V, Ansari JM, Jefferson KK, Cava F, Jacobs-Wagner C, Fikrig E (2017) Pathogen-mediated manipulation of arthropod microbiota to promote infection. Proc Natl Acad Sci U S A 114(5):E781–E790. https://doi.org/10.1073/pnas.1613422114
Ma M, Tu C, Luo J, Lu M, Zhang S, Xu L (2021) Metabolic and immunological effects of gut microbiota in leaf beetles at the local and systemic levels. Integr Zool 16(3):313–323. https://doi.org/10.1111/1749-4877.12528
Berasategui A, Shukla S, Salem H, Kaltenpoth M (2016) Potential applications of insect symbionts in biotechnology. Appl Microbiol Biotechnol 100(4):1567–1577. https://doi.org/10.1007/s00253-015-7186-9
Xie S, Lan Y, Sun C, Shao Y (2019) Insect microbial symbionts as a novel source for biotechnology. World J Microbiol Biotechnol 35(2):25. https://doi.org/10.1007/s11274-019-2599-8
Tikhe CV, Martin TM, Howells A, Delatte J, Husseneder C (2016) Assessment of genetically engineered Trabulsiella odontotermitis as a ‘Trojan Horse’ for paratransgenesis in termites. BMC Microbiol 16(1):202. https://doi.org/10.1186/s12866-016-0822-4
Zhao R, Han R, Qiu X, Yan X, Cao L, Liu X (2008) Cloning and heterologous expression of insecticidal-protein-encoding genes from Photorhabdus luminescens TT01 in Enterobacter cloacae for termite control. Appl Environ Microbiol 74(23):7219–7226. https://doi.org/10.1128/AEM.00977-08
Arora AK, Douglas AE (2017) Hype or opportunity? Using microbial symbionts in novel strategies for insect pest control. J Insect Physiol 103:10–17. https://doi.org/10.1016/j.jinsphys.2017.09.011
MsangoSoko K, Gandotra S, Chandel RK, Sharma K, Ramakrishinan B, Subramanian S (2020) Composition and diversity of gut bacteria associated with the eri silk moth, Samia ricini, (Lepidoptera: Saturniidae) as revealed by culture-dependent and metagenomics analysis. J Microbiol Biotechnol 30(9):1367–1378. https://doi.org/10.4014/jmb.2002.02055
Zhou F, Gao Y, Liu M, Xu L, Wu X, Zhao X, Zhang X (2021) Bacterial inhibition on Beauveria bassiana contributes to microbiota stability in Delia antiqua. Front Microbiol 12:710800. https://doi.org/10.3389/fmicb.2021.710800
Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, Bai Y, Bisanz JE, Bittinger K et al (2019) Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 37(8):852–857. https://doi.org/10.1038/s41587-019-0209-9
Zhao Y, Xue C-B, Yang L, Zhou C-G, Luo W-C (2010) Enzymatic dynamics of catechol oxidase from Gastrolina depressa. Pestic Biochem Physiol 96(2):57–62. https://doi.org/10.1016/j.pestbp.2009.09.007
Kutcherov D (2016) Temperature effects on the development, body size, and sex ratio of the walnut leaf beetle Gastrolina depressa (Coleoptera: Chrysomelidae). J Asia Pac Entomol 19(1):153–158. https://doi.org/10.1016/j.aspen.2016.01.002
Wang Q, Tang G (2018) The mitochondrial genomes of two walnut pests, Gastrolina depressa depressa and G depressa thoracica (Coleoptera: Chrysomelidae), and phylogenetic analyses. PeerJ 6:e4919. https://doi.org/10.7717/peerj.4919
Chang S-J, Park I-K (2011) Morphological and ecological study of Gastrolina depressa Baly (Coleoptera: Chrysomelidae). Korean journal of applied entomology 50(3):253–256. https://doi.org/10.5656/ksae.2011.08.0.37
Chang KS, Morimoto N (1988) Life table studies of the walnut leaf beetle, Gastrolina depressa (Coleoptera: Chrysomelidae), with special attention to aggregation. Res Popul Ecol 30(2):297–313. https://doi.org/10.1007/BF02513251
Kucuk RA (2020) Gut Bacteria in the holometabola: a review of obligate and facultative symbionts. J Insect Sci (Ludhiana) 20(4). https://doi.org/10.1093/jisesa/ieaa084
Salem H, Kirsch R, Pauchet Y, Berasategui A, Fukumori K, Moriyama M, Cripps M, Windsor D, Fukatsu T, Gerardo NM (2020) Symbiont Digestive Range Reflects Host Plant Breadth in Herbivorous Beetles. Curr Biol 30(15):2875-2886 e2874. https://doi.org/10.1016/j.cub.2020.05.043
Bozorov TA, Rasulov BA, Zhang D (2019) Characterization of the gut microbiota of invasive Agrilus mali Matsumara (Coleoptera: Buprestidae) using high-throughput sequencing: uncovering plant cell-wall degrading bacteria. Sci Rep 9(1):4923. https://doi.org/10.1038/s41598-019-41368-x
Berry D, Ben Mahfoudh K, Wagner M, Loy A (2011) Barcoded primers used in multiplex amplicon pyrosequencing bias amplification. Appl Environ Microbiol 77(21):7846–7849. https://doi.org/10.1128/AEM.05220-11
DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL (2006) Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72(7):5069–5072. https://doi.org/10.1128/AEM.03006-05
Magoc T, Salzberg SL (2011) FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27(21):2957–2963. https://doi.org/10.1093/bioinformatics/btr507
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D et al (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7(5):335–336. https://doi.org/10.1038/nmeth.f.303
Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP (2016) DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods 13(7):581–583. https://doi.org/10.1038/nmeth.3869
Hall M, Beiko RG (2018) 16S rRNA Gene Analysis with QIIME2. Methods in molecular biology (Clifton, N.J.) 1849: 113–129. https://doi.org/10.1007/978-1-4939-8728-3_8
Tettelin H, Masignani V, Cieslewicz MJ, Donati C, Medini D, Ward NL, Angiuoli SV, Crabtree J, Jones AL, Durkin AS, Deboy RT, Davidsen TM, Mora M et al (2005) Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome.” Proc Natl Acad Sci USA 102(39):13950–13955. https://doi.org/10.1073/pnas.0506758102
Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin P, O’Hara RB, Simpson G, Solymos P, Stevens MHH, Wagner H (2015) vegan: Community Ecology Package. R package version 2.2–1.
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG et al (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75(23):7537–7541. https://doi.org/10.1128/AEM.01541-09
Chao A, Bunge J (2002) Estimating the number of species in a stochastic abundance model. Biometrics 58(3):531–539. https://doi.org/10.1111/j.0006-341x.2002.00531.x
Chapman MG, Underwood AJ (1999) Ecological patterns in multivariate assemblages:information and interpretation of negative values in ANOSIM tests. Mar Ecol Prog Ser 180:257–265. https://doi.org/10.3354/meps180257
Bray JR, Curtis JT (1957) An ordination of the upland forest communities of Southern Wisconsin. Ecol Monogr 27(4). https://doi.org/10.2307/1942268
Warnes G, Bolker B, Bonebakker L, Gentleman R, Huber W, Liaw A, Lumley T, Mächler M, Magnusson A, Möller S. (2005). gplots: Various R programming tools for plotting data (Vol. 2).
Lex A, Gehlenborg N, Strobelt H, Vuillemot R, Pfister H (2014) UpSet: Visualization of Intersecting Sets. IEEE Trans Vis Comput Graph 20(12):1983–1992. https://doi.org/10.1109/TVCG.2014.2346248
Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C (2011) Metagenomic biomarker discovery and explanation. Genome Biol 12(6):R60. https://doi.org/10.1186/gb-2011-12-6-r60
Douglas GM, Beiko RG, Langille MGI (2018) Predicting the functional potential of the microbiome from marker genes using PICRUSt. Methods Molec Biol 1849:169–177. https://doi.org/10.1007/978-1-4939-8728-3_11
Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Vega Thurber RL, Knight R, Beiko RG, Huttenhower C (2013) Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 31(9):814–821. https://doi.org/10.1038/nbt.2676
Douglas GM, Maffei VJ, Zaneveld JR, Yurgel SN, Brown JR, Taylor CM, Huttenhower C, Langille MGI (2020) PICRUSt2 for prediction of metagenome functions. Nat Biotechnol 38(6):685–688. https://doi.org/10.1038/s41587-020-0548-6
Jang S, Kikuchi Y (2020) Impact of the insect gut microbiota on ecology, evolution, and industry. Curr Opin Insect Sci 41:33–39. https://doi.org/10.1016/j.cois.2020.06.004
Xue H, Zhu X, Wang L, Zhang K, Li D, Ji J, Niu L, Wu C, Gao X, Luo J, Cui J (2021) Gut Bacterial Diversity in Different Life Cycle Stages of Adelphocoris suturalis (Hemiptera: Miridae). Front Microbiol 12:670383. https://doi.org/10.3389/fmicb.2021.670383
Wang X, Sun S, Yang X, Cheng J, Wei H, Li Z, Michaud JP, Liu X (2020) Variability of gut microbiota across the life cycle of Grapholita molesta (Lepidoptera: Tortricidae). Front Microbiol 11:1366. https://doi.org/10.3389/fmicb.2020.01366
Wang M, Xiang X, Wan X (2020) Divergence in gut bacterial community among life stages of the rainbow stag beetle Phalacrognathus muelleri (Coleoptera: Lucanidae). Insects 11(10). https://doi.org/10.3390/insects11100719
Huang S, Zhang H (2013) The impact of environmental heterogeneity and life stage on the hindgut microbiota of Holotrichia parallela larvae (Coleoptera: Scarabaeidae). PLoS ONE 8(2):e57169. https://doi.org/10.1371/journal.pone.0057169
de Vries EJ, Jacobs G, Breeuwer JA (2001) Growth and transmission of gut bacteria in the Western flower thrips, Frankliniella occidentalis. J Invertebr Pathol 77(2):129–137. https://doi.org/10.1006/jipa.2001.5010
Shibl AA, Isaac A, Ochsenkuhn MA, Cardenas A, Fei C, Behringer G, Arnoux M, Drou N, Santos MP, Gunsalus KC, Voolstra CR, Amin SA (2020) Diatom modulation of select bacteria through use of two unique secondary metabolites. Proc Natl Acad Sci U S A 117(44):27445–27455. https://doi.org/10.1073/pnas.2012088117
Yin C, Sun F, Rao Q, Zhang Y (2020) Chemical compositions and antimicrobial activities of the essential oil from Pterocarya stenoptera C. DC Natural Product Research 34(19):2828–2831. https://doi.org/10.1080/14786419.2019.1587426
Dillon RJ, Dillon VM (2004) The gut bacteria of insects: nonpathogenic interactions. Annu Rev Entomol 49:71–92. https://doi.org/10.1146/annurev.ento.49.061802.123416
Chen B, Teh BS, Sun C, Hu S, Lu X, Boland W, Shao Y (2016) Biodiversity and activity of the gut microbiota across the life history of the insect herbivore Spodoptera littoralis. Sci Rep 6:29505. https://doi.org/10.1038/srep29505
Zhao C, Zhao H, Zhang S, Luo J, Zhu X, Wang L, Zhao P, Hua H, Cui J (2019) The developmental stage symbionts of the pea aphid-feeding Chrysoperla sinica (Tjeder). Front Microbiol 10:2454. https://doi.org/10.3389/fmicb.2019.02454
Ge SX, Shi FM, Pei JH, Hou ZH, Zong SX, Ren LL (2021) Gut bacteria associated with monochamus saltuarius (Coleoptera: Cerambycidae) and Their possible roles in host plant adaptations. Front Microbiol 12:687211. https://doi.org/10.3389/fmicb.2021.687211
Jones RT, Sanchez LG, Fierer N (2013) A cross-taxon analysis of insect-associated bacterial diversity. PLoS ONE 8(4):e61218. https://doi.org/10.1371/journal.pone.0061218
Yun JH, Roh SW, Whon TW, Jung MJ, Kim MS, Park DS, Yoon C, Nam YD, Kim YJ, Choi JH, Kim JY, Shin NR, Kim SH et al (2014) Insect gut bacterial diversity determined by environmental habitat, diet, developmental stage, and phylogeny of host. Appl Environ Microbiol 80(17):5254–5264. https://doi.org/10.1128/AEM.01226-14
Wang H, Cai W-M, Wang W-X, Yang J-M (2006) Molluscicidal activity of Nerium indicum Mill, Pterocarya stenoptera DC, and Rumex japonicum houtt on Oncomelania hupensis. Biomed Environ Sci 19(4):245–248
Delaviz H, Mohammadi J, Ghalamfarsa G, Mohammadi B, Farhadi N (2017) A review study on phytochemistry and pharmacology applications of Juglans Regia Plant. Pharmacogn Rev 11(22):145–152. https://doi.org/10.4103/phrev.phrev_10_17
Schwindl S, Kraus B, Heilmann J (2019) Secondary metabolites from the leaves of Juglans regia L. Biochem Syst Ecol 83:130–136. https://doi.org/10.1016/j.bse.2019.01.014
Abbott DW, Boraston AB (2008) Structural biology of pectin degradation by Enterobacteriaceae. Microbiol Mol Biol Rev 72(2):301–316. https://doi.org/10.1128/MMBR.00038-07
Xia X, Gurr GM, Vasseur L, Zheng D, Zhong H, Qin B, Lin J, Wang Y, Song F, Li Y, Lin H, You M (2017) Metagenomic sequencing of diamondback moth gut microbiome unveils key holobiont adaptations for herbivory. Front Microbiol 8:663. https://doi.org/10.3389/fmicb.2017.00663
Mason CJ, Jones AG, Felton GW (2019) Co-option of microbial associates by insects and their impact on plant-folivore interactions. Plant Cell Environ 42(3):1078–1086. https://doi.org/10.1111/pce.13430
Behar A, Yuval B, Jurkevitch E (2005) Enterobacteria-mediated nitrogen fixation in natural populations of the fruit fly Ceratitis capitata. Mol Ecol 14(9):2637–2643. https://doi.org/10.1111/j.1365-294X.2005.02615.x
Augustinos AA, Kyritsis GA, Papadopoulos NT, Abd-Alla AM, Caceres C, Bourtzis K (2015) Exploitation of the medfly gut microbiota for the enhancement of sterile insect technique: use of Enterobacter sp Larval Diet-Based Probiotic Applications. PLoS One 10(9):e0136459. https://doi.org/10.1371/journal.pone.0136459
Anand AA, Vennison SJ, Sankar SG, Prabhu DI, Vasan PT, Raghuraman T, Geoffrey CJ, Vendan SE (2010) Isolation and characterization of bacteria from the gut of Bombyx mori that degrade cellulose, xylan, pectin and starch and their impact on digestion. J Insect Sci (Ludhiana) 10:107. https://doi.org/10.1673/031.010.10701
Gonzalez-Serrano F, Perez-Cobas AE, Rosas T, Baixeras J, Latorre A, Moya A (2020) The gut microbiota composition of the moth brithys crini reflects insect metamorphosis. Microb Ecol 79(4):960–970. https://doi.org/10.1007/s00248-019-01460-1
Koskinioti P, Ras E, Augustinos AA, Tsiamis G, Beukeboom LW, Caceres C, Bourtzis K (2019) The effects of geographic origin and antibiotic treatment on the gut symbiotic communities of Bactrocera oleae populations. Entomol Exp Appl 167(3):197–208. https://doi.org/10.1111/eea.12764
Laviad-Shitrit S, Izhaki I, Whitman WB, Shapiro N, Woyke T, Kyrpides NC, Halpern M (2020) Draft genome of Rosenbergiella nectarea strain 8N4(T) provides insights into the potential role of this species in its plant host. PeerJ 8:e8822. https://doi.org/10.7717/peerj.8822
Briones-Roblero CI, Rodriguez-Diaz R, Santiago-Cruz JA, Zuniga G, Rivera-Orduna FN (2017) Degradation capacities of bacteria and yeasts isolated from the gut of Dendroctonus rhizophagus (Curculionidae: Scolytinae). Folia Microbiol (Praha) 62(1):1–9. https://doi.org/10.1007/s12223-016-0469-4
Setiaji J, Feliatra F, Teruna HY, Lukistyowati I, Suharman I, Muchlisin ZA, Johan TI (2020) Antibacterial activity in secondary metabolite extracts of heterotrophic bacteria against Vibrio alginolyticus, Aeromonas hydrophila, and Pseudomonas aeruginosa. F1000Res 9: 1491. https://doi.org/10.12688/f1000research.26215.1
Kim JM, Choi MY, Kim JW, Lee SA, Ahn JH, Song J, Kim SH, Weon HY (2017) Effects of diet type, developmental stage, and gut compartment in the gut bacterial communities of two Cerambycidae species (Coleoptera). J Microbiol 55(1):21–30. https://doi.org/10.1007/s12275-017-6561-x
Funding
This research was supported by the National Natural Science Foundation of China (31971663) and the Young Elite Scientists Sponsorship Program by CAST (2020QNRC001).
Author information
Authors and Affiliations
Contributions
Letian Xu and Meiqi Ma designed the whole experiment. Meiqi Ma, Xiaotong Chen, and Siqun Li collected the samples and conducted all experiments. Meiqi Ma, Letian Xu, and Jing Luo analyzed the data and wrote the manuscript, and Runhua Han, Xiaotong Chen, Jing Luo, and Letian Xu revised and submitted the manuscript.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare no competing interests.
Additional information
Meiqi Ma and Xiaotong Chen contribute equally to the study.
Supplementary Information
Below is the link to the electronic supplementary material.
248_2022_2054_MOESM1_ESM.pdf
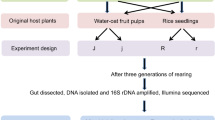
Supplementary file1 Sample collection of G. depressa and 16S rRNA sequencing process. The whole bodies of first and second instar larvae were sampled due to their small size and beetles in other life stages were dissected to collect guts. Each sequencing sample contained three whole bodies or three guts. L1, 1st instar larvae; L2, 2nd instar larvae; L3, 3rd instar larvae; Ma, male adults; Fe, female adults; Pre, pre-pregnant female adults. (PDF 1119 KB)
248_2022_2054_MOESM2_ESM.pdf
Supplementary file2 Pan/Core species analysis at the ASV level to assess whether sequenced samples were enough. (a) Pan species. (b) Core species. L1, 1st instar larvae; L2, 2nd instar larvae; L3, 3rd instar larvae; Ma, male adults; Fe, female adults; Pre, pre-pregnant female adults. (PDF 461 KB)
248_2022_2054_MOESM5_ESM.xls
Supplementary file5 The statistical table of difference by Kruskal-Wallis test with the Mann-Whitney U post hoc test, P < 0.05. (XLS 34 KB)
Rights and permissions
About this article
Cite this article
Ma, M., Chen, X., Li, S. et al. Composition and Diversity of Gut Bacterial Community in Different Life Stages of a Leaf Beetle Gastrolina depressa. Microb Ecol 86, 590–600 (2023). https://doi.org/10.1007/s00248-022-02054-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-022-02054-0