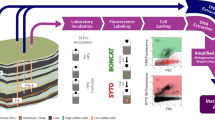
Abstract
The global trend of transiting to more renewable energy sources requires transition fuels, such as coal seam gas, to supplement and secure energy needs. In order to optimise strategies and technologies for enhancing gas production, an understanding of the fundamental microbial processes and interactions would be advantageous. Models have recently begun mapping the microbial roles and interactions in coal seam environments, from direct coal degradation to methanogenesis. This study seeks to expand those models by observing community compositional shifts in the presence of differing organic matter by conducting 16S rRNA microbial surveys using formation water from the Surat and Sydney Basins grown on varying types of organic matter (black and brown coal, oil shale, humic acid, and lignin). A total of 135 microbes were observed to become enriched in the presence of added organic matter in comparison to carbon-free treatments. These surveys allowed detailed analysis of microbial compositions in order to extrapolate which taxa favour growth in the presence of differing organic matter. This study has experimentally demonstrated shifts in the microbial community composition due to differing carbon sources and, for the first time, generated a conceptual model to map putative degradation pathways regarding subsurface microbial consortia.



Similar content being viewed by others
References
Barnhart EP, De León KB, Ramsay BD, Cunningham AB, Fields MW (2013) Investigation of coal-associated bacterial and archaeal populations from a diffusive microbial sampler (DMS). Int J Coal Geol 115:64–70
Beckmann S, Luk AWS, Gutierrez-Zamora ML, Chong NHH, Thomas T, Lee M, Manefield M (2019)Long-term succession in a coal seam microbiome during in situ biostimulation of coalbed-methane generation. ISME J 13:632–650
Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, Gormley N, Gilbert JA, Smith G, Knight R (2012)Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 6:1621–1624
Chew YM, Lye S, Salleh MM, Yahya A (2018) 16S rRNA metagenomic analysis of the symbiotic community structures of bacteria in foregut, midgut, and hindgut of the wood-feeding termite Bulbitermes sp. Symbiosis 76:187–197
Colberg PJ, Young LY (1985) Aromatic and volatile acid intermediates observed during anaerobic metabolism of lignin-derived oligomers. Appl Environ Microbiol 49:350–358
Cole JR, Wang Q, Fish JA, Chai B, McGarrell DM, Sun Y, Brown CT, Porras-Alfaro A, Kuske CR, Tiedje JM (2014) Ribosomal database project: data and tools for high throughput rRNA analysis. Nucleic Acids Res 42:D633–D642
Cowell, R.K. (2006) EstimateS: statistical estimation of species richness and shared species from simples, version 9.1.0. http://purl.oclc.org/estimates.
Di Gioia D, Barberio C, Spagnesi S, Marchetti L, Fava F (2002) Characterization of four olive-mill-wastewater indigenous bacterial strains capable of aerobically degrading hydroxylated and methoxylated monocyclic aromatic compounds. Arch Microbiol 178:208–217
Edgar RC (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461
Gibson J, Harwood CS (1995) Degradation of aromatic compounds by nonsulfur purple bacteria. In: Blankenship RE, Madigan MT, Bauer CE (eds)Anoxygenic Photosynthetic Bacteria. Springer, Dordrecht, pp 991–1003
Gößner A, Daniel SL, Drake HL (1994) Acetogenesis coupled to the oxidation of aromatic aldehyde groups. Arch Microbiol 161:126–131
Green MS, Flanegan KC, Gilcrease PC (2008) Characterization of a methanogenic consortium enriched from a coalbed methane well in the Powder River Basin, USA. Int J Coal Geol 76:34–45
Halliday MJ, Giles HE, Padmanabha J, McSweeney CS, Dalzell SA, Max Shelton H (2018) The efficacy of a cultured Synergistes jonesii inoculum to control hydroxypyridone toxicity in Bos indicus steers fed leucaena/grass diets. Anim Produc Sci 59:696–708
Hammer Ø, Harper DAT, Ryan PD, Others (2001) PAST: Paleontological statistics software package for education and data analysis. Palaeontol Electronica 4:9
Hatcher PG, Clifford DJ (1997) The organic geochemistry of coal: from plant materials to coal. Org Geochem 27:251–274
Hug LA, Baker BJ, Anantharaman K, Brown CT, Probst AJ, Castelle CJ, Butterfield CN, Hernsdorf AW, Amano Y, Ise K, Suzuki Y, Dudek N, Relman DA, Finstad KM, Amundson R, Thomas BC, Banfield JF (2016) A new view of the tree of life. Nat Microbiol 1:1–6
Hunter JD (2007) Matplotlib: A 2D Graphics Environment. Comput Sci Eng 9:90–95
Janusz G, Pawlik A, Sulej J, Swiderska-Burek U, Jarosz-Wilkolazka A, Paszczynski A (2017) Lignin degradation: microorganisms, enzymes involved, genomes analysis and evolution. FEMS Microbiol Rev 41:941–962
Kappell AD, Wei Y, Newton RJ, Van Nostrand JD, Zhou J, McLellan SL, Hristova KR (2014) The polycyclic aromatic hydrocarbon degradation potential of Gulf of Mexico native coastal microbial communities after the Deepwater Horizon oil spill. Front Microbiol 5:205
Kato S, Chino K, Kamimura N, Masai E, Yumoto I, Kamagata Y (2015) Methanogenic degradation of lignin-derived monoaromatic compounds by microbial enrichments from rice paddy field soil. Sci Rep 5:1–11
Khan MU, Ahring BK (2019) Lignin degradation under anaerobic digestion: Influence of lignin modifications -a review. Biomass Bioenergy 128:105325
Lal B, Khanna S (1996) Degradation of crude oil by Acinetobacter calcoaceticus and Alcaligenes odorans. J Appl Bacteriol 81:355–362
Larsen SB, Karakashev D, Angelidaki I, Schmidt JE (2009)Ex-situ bioremediation of polycyclic aromatic hydrocarbons in sewage sludge. J Hazard Mater 164:1568–1572
Matsushita M, Magara K, Sato Y, Shinzato N, Kimura H (2018) Geochemical and microbiological evidence for microbial methane production in deep aquifers of the cretaceous accretionary prism. Microbes Environ 33:205–213
Mayumi D, Mochimaru H, Tamaki H, Yamamoto K, Yoshioka H, Suzuki Y, Kamagata Y, Sakata S (2016) Methane production from coal by a single methanogen. Science 354:222–225
Mazzoli R, Pessione E, Giuffrida MG, Fattori P, Barello C, Giunta C, Lindley ND (2007) Degradation of aromatic compounds by Acinetobacter radioresistens S13: growth characteristics on single substrates and mixtures. Arch Microbiol 188:55–68
Meslé M, Périot C, Dromart G, Oger P (2013) Biostimulation to identify microbial communities involved in methane generation in shallow, kerogen-rich shales. J Appl Microbiol 114:55–70
Meslé M, Dromart G, Haeseler F, Oger PM (2015) Classes of organic molecules targeted by a methanogenic microbial consortium grown on sedimentary rocks of various maturities. Front Microbiol6:589
Midgley DJ, Hendry P, Pinetown KL, Fuentes D, Gong S, Mitchell DL, Faiz M (2010) Characterisation of a microbial community associated with a deep, coal seam methane reservoir in the Gippsland Basin, Australia. Int J Coal Geol 82:232–239
Musat F, Galushko A, Jacob J, Widdel F, Kube M, Reinhardt R, Wilkes H, Schink B, Rabus R (2009) Anaerobic degradation of naphthalene and 2-methylnaphthalene by strains of marine sulfate-reducing bacteria. Environ Microbiol 11:209–219
Noguchi M, Kurisu F, Kasuga I, Furumai H (2014)Time-resolved DNA stable isotope probing links Desulfobacterales- and Coriobacteriaceae-related bacteria to anaerobic degradation of benzene under methanogenic conditions. Microbes Environ 29:191–199
Ransley T, Radke BM, Carey H, Stewart G, Bell J, Owens R et al (2015) Hydrogeological Atlas of the Great Artesian Basin. Geoscience Australia, pp 134
Rice DD, Claypool GE (1981) Generation, accumulation, and resource potential of biogenic gas. AAPG Bull 65:5–25
Ritter D, Vinson D, Barnhart E, Akob DM, Fields MW, Cunningham AB, Orem W, McIntosh JC (2015) Enhanced microbial coalbed methane generation: a review of research, commercial activity, and remaining challenges. Int J Coal Geol 146:28–41
RStudio Team (2016) RStudio: integrated development environment for R (Version 1.4.1103) [Computer software]. Rstudio, Boston, MA
Stout SA, Spackman W (1987) A microscopic investigation of woody tissues in peats: Some processes active in the peatification of Ligno-cellulosic cell walls. Int J Coal Geol 8:55–68
Strąpoć D, Picardal FW, Turich C, Schaperdoth I, Macalady JL, Lipp JS et al (2008)Methane-Producing Microbial Community in a Coal Bed of the Illinois Basin. Appl Environ Microbiol 74:2424–2432
Strąpoć D, Mastalerz M, Dawson K, Macalady J, Callaghan AV, Wawrik B, Turich C, Ashby M (2011) Biogeochemistry of Microbial Coal-Bed Methane. Annu Rev Earth Planet Sci 39:617–656
Ueno A, Shimizu S, Tamamura S, Okuyama H, Naganuma T, Kaneko K (2016) Anaerobic decomposition of humic substances by Clostridium from the deep subsurface. Sci Rep 6:18990
van der Walt S, Colbert SC, Varoquaux G (2011) The NumPy array: a structure for efficient numerical computation. Comput Sci Eng 13:22–30
Vick SHW, Tetu SG, Sherwood N, Pinetown K, Sestak S, Vallotton P, Elbourne LDH, Greenfield P, Johnson E, Barton D, Midgley DJ, Paulsen IT (2016) Revealing colonisation and biofilm formation of an adherent coal seam associated microbial community on a coal surface. Int J Coal Geol 160-161:42–50
Vick SHW, Greenfield P, Tran-Dinh N, Tetu SG, Midgley DJ, Paulsen IT (2018) The Coal Seam Microbiome (CSMB) reference set, a lingua franca for the microbial coal-to-methane community. Int J Coal Geol 186:41–50
Vick SHW, Gong S, Sestak S, Vergara TJ, Pinetown KL, Li Z et al (2019) Who eats what? Unravelling microbial conversion of coal to methane. FEMS Microbiol Ecol 95:fiz093
Vick SHW, Greenfield P, Pinetown KL, Sherwood N, Gong S, Tetu SG et al (2019) Succession patterns and physical niche partitioning in microbial communities from subsurface coal seams. iScience 12:152–167
Vick SHW, Greenfield P, Tetu SG, Midgley DJ, Paulsen IT (2019) Genomic and phenotypic insights point to diverse ecological strategies by facultative anaerobes obtained from subsurface coal seams. Sci Rep 9:16186
Widdel F, Kohring GW, Mayer F (1983) Studies on dissimilatory sulfate-reducing bacteria that decompose fatty acids: III. Characterization of the filamentous gliding Desulfonema limicola gen. nov. sp. nov., and Desulfonema magnum sp. nov. Arch Microbiol 134:286–294
Wolin EA, Wolin MJ, Wolfe RS (1963) Formation of methane by bacterial extracts. J Biol Chem 238:2882–2886
Xu B, Xu W, Yang F, Li J, Yang Y, Tang X, Mu Y, Zhou J, Huang Z (2013) Metagenomic analysis of the pygmy loris fecal microbiome reveals unique functional capacity related to metabolism of aromatic compounds. PLoS One 8:e56565
Young LY, Frazer AC (1987) The fate of lignin and lignin-derived compounds in anaerobic environments. Geomicrobiol J 5:261–293
Zealand AM, Mei R, Papachristodoulou P, Roskilly AP, Liu WT, Graham DW (2018) Microbial community composition and diversity in rice straw digestion bioreactors with and without dairy manure. Appl Microbiol Biotechnol 102:8599–8612
Acknowledgements
The authors thank Dr. Neil Sherwood and Dr. Zhongsheng Li for the photos and petrological analyses and Dr. Phil Hendry for the assistance with formulation of the chemical medium. Thanks also to Macrogen (Korea) for DNA sequencing. The authors also thank Origin and AGL for the access to their coal seam gas production areas for sampling of formation waters. The gamma irradiation in the present study was conducted with support from Professors Robert Willows and Jan Gebicki for which we are grateful. We also thank Ms. Bronwyn Campbell for the useful discussions on these processes and their relationships to geology. Mr. Andrew McLeish is supported through a postgraduate scholarship from Macquarie University and additional scholarship support from the Commonwealth Scientific and Industrial Research Organisation, Energy.
Availability of data and material
All OTU sequences were submitted to GenBank under the accession numbers MN929133-MN929716.
Code availability
Not applicable.
Funding
This study was supported through a postgraduate scholarship from Macquarie University and additional scholarship support from the Commonwealth Scientific and Industrial Research Organisation, Energy.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
The authors give consent for publication.
Conflict of interest
The authors declare no competing interests.
Rights and permissions
About this article
Cite this article
McLeish, A.G., Gong, S., Greenfield, P. et al. Microbial Community Shifts on Organic Rocks of Different Maturities Reveal potential Catabolisers of Organic Matter in Coal. Microb Ecol 84, 780–793 (2022). https://doi.org/10.1007/s00248-021-01857-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-021-01857-x