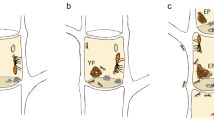
Abstract
Understanding the ecological processes that shape species assemblage patterns is central to community ecology. The effects of ecological processes on assemblage patterns are scale-dependent. We used metabarcoding and shotgun sequencing to determine bacterial taxonomic and functional assemblage patterns among varying defined focal scales (micro-, meso-, and macroscale) within the American alligator (Alligator mississippiensis) nesting microbiome. We correlate bacterial assemblage patterns among eight nesting compartments within and proximal to alligator nests (micro-), across 18 nests (meso-), and between 4 geographic sampling sites (macro-), to determine which ecological processes may drive bacterial assemblage patterns within the nesting environment. Among all focal scales, bacterial taxonomic and functional richness (α-diversity) did not statistically differ. In contrast, bacterial assemblage structure (β-diversity) was unique across all focal scales, whereas functional pathways were redundant within nests and across geographic sites. Considering these observed scale-based patterns, taxonomic bacterial composition may be governed by unique environmental filters and dispersal limitations relative to microbial functional attributes within the alligator nesting environment. These results advance pattern-process dynamics within the field of microbial community ecology and describe processes influencing the American alligator nest microbiome.






Similar content being viewed by others
References
Vellend M (2010) Conceptual synthesis in community ecology. Q Rev Biol 85:183–206. https://doi.org/10.1086/652373
Sydenham MAK et al (2017) Disentangling the contributions of dispersal limitation, ecological drift, and ecological filtering to wild bee community assembly. Ecos 8:e01650. https://doi.org/10.1002/ecs2.1650
MacArthur RH (1969) Patterns of communities in the tropics. Biol J Linn Soc 1:19–30. https://doi.org/10.1111/j.1095-8312.1969.tb01809.x
Ron R, Fragman-Sapir O, Kadmon R (2018) Dispersal increases ecological selection by increasing effective community size. Proc Natl Acad Sci U S A 115:11280–11285. https://doi.org/10.1073/pnas.1812511115
Bowler DE, Benton TG (2005) Causes and consequences of animal dispersal strategies: relating individual behaviour to spatial dynamics. Biol Rev Camb Philos Soc 80:205–225. https://doi.org/10.1017/s1464793104006645
Schluter D, Conte GL (2009) Genetics and ecological speciation. Proc Natl Acad Sci U S A 106:9955–9962. https://doi.org/10.1073/pnas.0901264106
Gilbert B, Levine JM (2017) Ecological drift and the distribution of species diversity. Proc Biol Sci 284:20170507. https://doi.org/10.1098/rspb.2017.0507
Renault D, Laparie M, McCauley SJ, Bonte D (2018) Environmental adaptations, ecological filtering, and dispersal central to insect invasions. Annu Rev Entomol 63:345–368. https://doi.org/10.1146/annurev-ento-020117-043315
Kraft NJB et al (2015) Community assembly, coexistence and the environmental filtering metaphor. Funct Ecol 29:592–599. https://doi.org/10.1111/1365-2435.12345
Wiens JA (1989) Spatial scaling in ecology. Funct Ecol 3:385–397. https://doi.org/10.2307/2389612
Levin SA (1995) The problem of pattern and scale in ecology. Ecology. 73:277–326. https://doi.org/10.2307/1941447
Belmaker J, Jetz W (2013) Spatial scaling of functional structure in bird and mammal assemblages. Am Nat 181:464–478. https://doi.org/10.1086/669906
Terlizzi A, Anderson MJ, Fraschetti S, Benedetti-Cecchi L (2007) Scales of spatial variation in Mediterranean subtidal sessile assemblages at different depths. Mar Ecol Prog Ser 332:25–39. https://doi.org/10.3354/meps332025
Boon E et al (2013) Interactions in the microbiome: communities of organisms and communities of genes. FEMS Microbiol Rev 38:90–118. https://doi.org/10.1111/1574-6976.12035
Nemergut DR et al (2013) Patterns and processes of microbial community assembly. Microbiol Mol Biol Rev 77:342–356. https://doi.org/10.1128/MMBR.00051-12
Morrison-Whittle P, Goddard MR (2015) Quantifying the relative roles of selective and neutral processes in defining eukaryotic microbial communities. ISME J 9:2003–2011. https://doi.org/10.1038/ismej.2015.18
Baxter AM, Johnson L, Edgerton J, Royer T, Leff LG (2012) Structure and function of denitrifying bacterial assemblages in low-order Indian streams. Fresh Sci 31:304–317. https://doi.org/10.1899/11-066.1
Branco S, Bruns TD, Singleton I (2013) Fungi at a small scale: spatial zonation of fungal assemblages around single trees. PLoS ONE 8:e78295. https://doi.org/10.1371/journal.pone.0078295
Canfora L et al (2014) Salinity and bacterial diversity: to what extent does the concentration of salt affect the bacterial community in a saline soil? PLoS ONE 9:e106662. https://doi.org/10.1371/journal.pone.0106662
Gourmelon V et al (2016) Environmental and geographical factors structure soil microbial diversity in new caledonian ultramafic substrates: a metagenomic approach. PLoS ONE 11:e0167405. https://doi.org/10.1371/journal.pone.0167405,
Caporaso JG et al (2011) Moving pictures of the human microbiome. Genome Biol 12:R50. https://doi.org/10.1186/gb-2011-12-5-r50
Colston TJ, Jackson CR (2016) Microbiome evolution along divergent branches of the vertebrate tree of life: what is known and unknown. Mol Ecol 25:3776–3800. https://doi.org/10.1111/mec.13730
Gilbert JA et al (2018) Current understanding of the human microbiome. Nat Med 24:394–400. https://doi.org/10.1038/nm.4517
Turnbaugh PJ et al (2007) The human microbiome project: exploring the microbial part of ourselves in a changing world. Nature. 449:804–810. https://doi.org/10.1038/nature06244
Burke C, Steinberg P, Rusch D, Kjelleberg S, Thomas T (2011) Bacterial community assembly based on functional genes rather than species. Proc Natl Acad Sci U S A 108:14288–14293. https://doi.org/10.1073/pnas.1101591108
Louca S et al (2016) High taxonomic variability despite stable functional structure across microbial communities. Nat Ecol Evol 1:0015. https://doi.org/10.1038/s41559-016-0015
Louca S et al (2018) Function and functional redundancy in microbial systems. Nat Ecol Evol 2:936–943. https://doi.org/10.1038/s41559-018-0519-1
Bletz MC et al (2016) Amphibian gut microbiota shifts differentially in community structure but converges on habitat specific predicted functions. Nat Commun 7:13699. https://doi.org/10.1038/ncomms13699
Jauker FT, Diekötter T, Schwarzbach F, Wolters V (2009) Pollinator dispersal in an agricultural matrix: opposing responses to landscape structure and distance from main habitat. Landsc Ecol 24:547–555. https://doi.org/10.1007/s10980-009-9331-2
Peay KG, Garbelotto M, Bruns TD (2010) Evidence of dispersal limitation in soil microorganisms: isolation reduces species richness on mycorrhizal tree islands. Ecology. 91:3631–3640. https://doi.org/10.1890/09-2237.1
Welsh RM et al (2016) Bacterial predation in a marine host-associated microbiome. ISME J 10:1540–1544. https://doi.org/10.1038/ismej.2015.219
Harris RN, James TY, Lauer A, Alice Simon A, Patel A (2006) Amphibian pathogen Batrachochytrium dendrobatidis is inhibited by the cutaneous bacteria of amphibian species. EcoHealth. 3:53–56. https://doi.org/10.1007/s10393-005-0009-1
Sharon G, Segal D, Zilber-Rosenberg I, Rosenberg E (2011) Symbiotic bacteria are responsible for diet-induced mating preference in Drosophila melanogaster, providing support for the hologenome concept of evolution. Gut Microbes 2:190–192. https://doi.org/10.4161/gmic.2.3.16103
Jacob S et al (2015) Microbiome affects egg carotenoid investment, nestling development and adult oxidative costs of reproduction in great tits. Funct Ecol 29:1048–1058. https://doi.org/10.1111/1365-2435.12404
Kearns PJ et al (2017) Fight fungi with fungi: antifungal properties of the amphibian mycobiome. Front Microbiol 8:2494. https://doi.org/10.3389/fmicb.2017.02494
Wiley NC et al (2017) The microbiota-gut-brain axis as a key regulator of neural function and the stress response: implications for human and animal health. J Anim Sci 95:3225–3246. https://doi.org/10.2527/jas.2016.1256
Kwak MJ et al (2018) Rhizosphere microbiome structure alters to enable wilt resistance in tomato. Nat Biotechnol 36:1100–1109. https://doi.org/10.1038/nbt.4232
Flechas SV et al (2019) Microbiota and skin defense peptides may facilitate coexistence of two sympatric Andean frog species with a lethal pathogen. ISME J 13:361–373. https://doi.org/10.1038/s41396-018-0284-9
Walker DM, Leys JE, Grisnik M, Grajal-Puche A, Murray CM, Allender MC (2019) Variability in snake skin microbial assemblages across spatial scales and disease states. ISME J 13:2209–2222. https://doi.org/10.1038/s41396-019-0416-x
Ferguson MWJ (1981) Extrinsic microbial degradation of the alligator eggshell. Science. 214:1135–1137. https://doi.org/10.1126/science.214.4525.1135
Ferguson MWJ (1982) The structure and composition of the eggshell and embryonic membranes of Alligator mississippiensis. Trans Zool Soc Lond 36:99–152. https://doi.org/10.1111/j.1096-3642.1982.tb00064.x
Joanen T, McNease L (1989) Ecology and physiology of nesting and early development of the American alligator. Integr Comp Biol 29:987–998. https://doi.org/10.1093/icb/29.3.987
Palmer ML, Mazzotti FJ (2004) Structure of everglades alligator holes. Wetlands. 24:115–122. https://doi.org/10.1672/0277-5212(2004)024[0115:SOEAH]2.0.CO;2
Merchant M, Murray CM, Cooper A (2014) American alligator nests as microhabitats for a diversity of vertebrates. Herp Rev 45:201–203
Grajal A (2019) Nesting alligator. Seattle, Washington
Grajal-Puche A, Kearley M, Bravo CA, Murray CM (2018) Alligator mississippiensis (American alligator). unique nesting ecology. Herp Rev 49:734–736
Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD (2013) Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol 79:5112–5120. https://doi.org/10.1128/AEM.01043-13
Schloss PD et al (2009) Introducing mothur: Open-Source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. https://doi.org/10.1128/AEM.01541-09
Quast C et al (2013) The SILVA ribosomal rna gene database project: improved data processing and web-based tools. Nucl Acid Res 41:D590–D596. https://doi.org/10.1093/nar/gks1219/
Rognes T, Flouro T, Nichols B, Quince C, Mahe F (2016) VSEARCH: a versatile open source tool for metagenomics. PeerJ. 4:e2584. https://doi.org/10.7717/peerj.2584
Comeau AM, Douglas GM, Langille GIM (2017) Microbiome helper: a custom and streamlined workflow for microbiome research. mSystems 2:e00127–e00116. https://doi.org/10.1128/mSystems.00127-16
Segata N et al (2012) Metagenomic microbial community profiling using unique clade-specific marker genes. Nat Methods 9:811–814. https://doi.org/10.1038/nmeth.2066
Franzosa EA et al (2018) Species-level functional profiling of metagenomes and metatranscriptomes. Nat Methods 15:962–968. https://doi.org/10.1038/s41592-018-0176-y
R Core Team (2019) R: a language and environment for statistical computing. R Foundation for Statistical Computing. Available from: https://www.R-project.org/
Oksanen J et al (2019) vegan: Community ecology package. R package version 2.5-4, Available from https://CRAN.R-project.org/package=vegan
De Cáceres M, Legendre P (2009) Associations between species and groups of sites: indices and statistical inference. Ecology. 90:3566–3574. https://doi.org/10.1890/08-1823.1
Simpson EH (1949) Measurement of diversity. Nature. 163:688. https://doi.org/10.1038/163688a0
Chen KT, Tan J, Way, G. P.. DOIng, G., Deborah, H. A. & Greene, C. S. (2018) PatheCORE-T: identifying and visualizing globally co-occuring pathways in large transcriptomic compendia. BioData Mining 11. https://doi.org/10.1186/s13040-018-0175-7
Ngai JT, Srivastava DS (2006) Predators accelerate nutrient cycling in a bromeliad ecosystem. Science 314. https://doi.org/10.1126/science.1132598
Murray CM, Easter M, Merchant M, Cooper A, Crother BI (2013) Can reproductive allometry assess population marginality in crocodilians? a comparative analysis of gulf coast American alligator (Alligator mississippiensis) populations. Copeia. 2:268–276. https://doi.org/10.1643/CH-11-136
Finlay BJ, Clarke KJ (1999) Ubiquitous dispersal of microbial species. Nature. 400:828. https://doi.org/10.1038/23616
Darcy JL, Lynch RC, King AJ, Robeson MS, Schmidt SK (2011) Global distribution of polaromonas phylotypes–evidence for a highly successful dispersal capacity. PLoS ONE 6:e23742. https://doi.org/10.1371/journal.pone.0023742
Wilkinson DM, Koumoutsaris S, Mitchell EAD, Bey I (2012) Modelling the effect of size on the aerial dispersal of microorganisms. J Biogeogr 39:89–97. https://doi.org/10.1111/j.1365-2699.2011.02569.x
Svoboda P, Lindström ES, Ahmed Osman O, Langenheder S (2018) Dispersal timing determines the importance of priority effects in bacterial communities. ISME J 12:644–646. https://doi.org/10.1038/ismej.2017.180
Fukami T et al (2010) Assembly history dictates ecosystem functioning: evidence from wood decomposer communities. Ecol Lett 13:675–684. https://doi.org/10.1111/j.1461-0248.2010.01465.x
Murray CM, Easter M, Padilla S, Marin MS, Guyer C (2016) Regional warming and the thermal regimes of American crocodile nests in the Tempisque basin, Costa Rica. J Therm Biol 60:49–59. https://doi.org/10.1016/j.jtherbio.2016.06.004
Peralta AL, Ludmer S, Kent AD (2013) Hydrologic history influences microbial community composition and nitrogen cycling under experimental drying/wetting treatments. Soil Biol Biochem 66:29–37. https://doi.org/10.1016/j.soilbio.2013.06.019
Hu A et al (2019) Diurnal temperature variation and plants drive latitudinal patterns in seasonal dynamics of soil microbial community. Front Microbiol 10:00674. https://doi.org/10.3389/fmicb.2019.00674
Lear G, Bellamy J, Case BS, Lee JE, Buckley HL (2014) Fine-scale spatial patterns in bacterial community composition and function within freshwater ponds. ISME J 8:1715–1726. https://doi.org/10.1038/ismej.2014.21
Oberbeckmann S, Kreikemeyer B, Labrenz M (2018) Environmental factors support the formation of specific bacterial assemblages on microplastics. Front Microbiol 8:2709. https://doi.org/10.3389/fmicb.2017.02709
Telford RJ, Vandvik V, Birks HJB (2006) Dispersal limitations matter for microbial morphospecies. Science. 312:1015. https://doi.org/10.1126/science.1125669
Ma H, Su H (2019) Effect of temperature on the fermentation of starch by two high efficient H2 producers. Renew Energy 138:964–970. https://doi.org/10.1016/j.renene.2019.01.126
Schweitzer B, Huber I, Amann R, Ludwig W, Simon M (2001) Alpha- and beta-Proteobacteria control the consumption and release of amino acids on lake snow aggregates. Appl Environ Microbiol 67:632–645. https://doi.org/10.1128/AEM.67.2.632-645.2001
Wang J, Liu Z, Xia J, Chen Y (2019) Effect of microbial inoculation on physicochemical properties and bacterial community structure of citrus peel composting. Bioresour Technol 291:121843. https://doi.org/10.1016/j.biortech.2019.121843
Zhou Y-M, Chen Y-P, Guo J-S, Shen Y, Yang J-X (2019) The correlations and spatial characteristics of microbiome and silage quality by reusing of citrus waste in a family-scale bunker silo. J Clean Prod 226:407–418. https://doi.org/10.1016/j.jclepro.2019.04.075
Acknowledgments
We thank C. and D. Kearley for logistical support and A. Grajal for artwork design.
Availability of Data and Materials
All raw sequence data can be found under GenBank SRA accession number(s) PRJNA554418 (amplicon analysis) and PRJNA554694 (metagenomic analysis). Statistical analysis was performed in R version 3.5.1; the corresponding R markdown code file is included in the supplemental material.
Author information
Authors and Affiliations
Contributions
AGP, DMW, and CMM conceived the study. All authors contributed to collection of specimens during field work. AGP and DMW completed the high-throughput sequencing, ran the bioinformatics, and statistical analyses. AGP, DMW, and CMM wrote the manuscript and all authors contributed equally to revisions.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Ethics Approval
Tennessee Technological University research policies and guidelines for the ethical treatment of animals were followed during this study (TTU-IACUC—17-18—007). Research collection permits were obtained from the appropriate governmental organizations (Eufaula National Wildlife Refuge Permit #: 4356020181R1; Alabama Conservation License Permit #: 201807822268680; J.D. Murphree Wildlife Management Area Collection Permit #: TPWD ESCA No. 826). Samples from Cameron Parish, LA were collected from private land under owner consent.
Electronic Supplementary Materials
Supplementary Table I
Taxonomic classification to the rank of order for bacteria indicative of the eggshell microbiome and other within nest assemblages. Indicator value > 0.50, α < 0.01. Highlighted rows are enriched (>3000 reads) bacterial taxa found only on the eggshell surface. (PDF 51 kb)
Supplementary Table II
Functional pathways indicative (indicator value > 0.50, α < 0.01) of the nest chamber (sample E) which is in direct contact with eggshell surfaces. Bolded pathways bordered by “ ** ” designate substrate degredation pathways. (PDF 86 kb)
Supplementary Table III
GPS coordinates of nests sampled in 2018. (PDF 30 kb)
ESM 1
(CSV 120 kb)
ESM 2
(CSV 2063 kb)
ESM 3
(CSV 2063 kb)
Rights and permissions
About this article
Cite this article
Grajal-Puche, A., Murray, C.M., Kearley, M. et al. Microbial Assemblage Dynamics Within the American Alligator Nesting Ecosystem: a Comparative Approach Across Ecological Scales. Microb Ecol 80, 603–613 (2020). https://doi.org/10.1007/s00248-020-01522-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-020-01522-9