Abstract
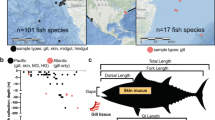
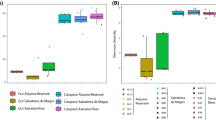
Microorganisms are an important component in shaping the evolution of hosts and as such, the study of bacterial communities with molecular techniques is shedding light on the complexity of symbioses between bacteria and vertebrates. Teleost fish are a heterogeneous group that live in a wide variety of habitats, and thus a good model group to investigate symbiotic interactions and their influence on host biology and ecology. Here we describe the microbiota of thirteen teleostean species sharing the same environment in the Mediterranean Sea and compare bacterial communities among different species and body sites (external mucus, skin, gills, and intestine). Our results show that Proteobacteria is the dominant phylum present in fish and water. However, the prevalence of other bacterial taxa differs between fish and the surrounding water. Significant differences in bacterial diversity are observed among fish species and body sites, with higher diversity found in the external mucus. No effect of sampling time nor species individual was found. The identification of indicator bacterial taxa further supports that each body site harbors its own characteristic bacterial community. These results improve current knowledge and understanding of symbiotic relationships among bacteria and their fish hosts in the wild since the majority of previous studies focused on captive individuals.



Similar content being viewed by others
References
Mcfall-Ngai M, Hadfield MG, Bosch TC et al (2013) Animals in a bacterial world, a new imperative for the life sciences. Proc. Natl. Acad. Sci. U. S. A. 110:3229–3236
Ghanbari M, Kneifel W, Domig KJ (2015) A new view of the fish gut microbiome: advances from next-generation sequencing. Aquaculture 448:464–475
Kohl KD (2018) A microbial perspective on the grand challenges in comparative animal physiology. mSystems 3:e00146-17
Nelson JS, Grande TC, Mark WVH (2016) Fishes of the world5th edn. John Wiley & Sons, New York
Ciric M, Waite D, Draper J, Jones JB (2018) Characterisation of gut microbiota of farmed Chinook salmon using metabarcoding. BioRxiv:288761 https://www.biorxiv.org/content/early/2018/03/26/288761.full.pdf+html
Barko PC, McMichael MA, Swanson KS, Williams DA (2018) The gastrointestinal microbiome: a review. J. Vet. Intern. Med. 32:9–25
Rosado D, Pérez-Losada M, Severino R, Cable J, Xavier R (2018) Characterization of the skin and gill microbiomes of the farmed seabass (Dicentrarchus labrax) and seabream (Sparus aurata). Aquaculture 500:57–64. https://doi.org/10.1016/j.aquaculture.2018.09.063
Tarnecki AM, Burgos FA, Ray CL, Arias CR (2017) Fish intestinal microbiome: diversity and symbiosis unraveled by metagenomics. J. Appl. Microbiol. https://doi.org/10.1111/jam.13415
Reverter M, Sasal P, Tapissier-Bontemps N, Lecchini D, Suzuki M (2017) Characterisation of the gill mucosal bacterial communities of four butterflyfish species: a reservoir of bacterial diversity in coral reef ecosystems. FEMS Microbiol. Ecol. 93(6). https://doi.org/10.1093/femsec/fix051
Llewellyn MS, Boutin S, Hoseinifar SH, Derome N (2014) Teleost microbiomes: the state of the art in their characterization, manipulation and importance in aquaculture and fisheries. Front. Microbiol. 5:207. https://doi.org/10.3389/fmicb.2014.00207/abstract
Sullam KE, Essinger SD, Lozupone CA, O’Connor MP, Rosen GL, Knight R, Kilham SS, Russell JA (2012) Environmental and ecological factors that shape the gut bacterial communities of fish: a meta-analysis. Mol. Ecol. 21:3363–3378
Eichmiller JJ, Hamilton MJ, Staley C, Sadowsky MJ, Sorensen PW (2016) Environment shapes the fecal microbiome of invasive carp species. Microbiome 4:44
Li X, Ringø E, Hoseinifar SH, Lauzon HL, Birkbeck H, Yang D (2018) The adherence and colonization of microorganisms in fish gastrointestinal tract. Rev Aquacult. https://doi.org/10.1111/raq.12248
Stagaman K, Burns AR, Guillemin K, Bohannan BJM (2017) The role of adaptive immunity as an ecological filter on the gut microbiota in zebrafish. ISME J 11:1630–1639
Wong S, Rawls JF (2012) Intestinal microbiota composition in fishes is influenced by host ecology and environment. Mol. Ecol. 21:3100–3102
Nayak SK (2010) Role of gastrointestinal microbiota in fish. Aquac. Res. 41:1553–1573
Xing M, Hou Z, Yuan J, Liu Y, Qu Y, Liu B (2013) Taxonomic and functional metagenomic profiling of gastrointestinal tract microbiome of the farmed adult turbot (Scophthalmus maximus). FEMS Microbiol. Ecol. 86:432–443
Li T, Long M, Gatesoupe FJ, Zhang Q, Li A, Gong X (2015) Comparative analysis of the intestinal bacterial communities in different species of carp by pyrosequencing. Microb. Ecol. 69:25–36
Ellis AE (2001) Innate host defense mechanisms of fish against. Dev. Comp. Immunol. 25:827–839
Chen CY, Chen PC, Weng FCH, Shaw GTW, Wang D (2017) Habitat and indigenous gut microbes contribute to the plasticity of gut microbiome in oriental river prawn during rapid environmental change. PLoS One 12:e0181427
Esteban MA (2012) An overview of the immunological defenses in fish skin. ISRN Immunol 853470:1–29. https://doi.org/10.5402/2012/853470
Legrand TPRA, Catalano SR, Wos-Oxley ML, Stephens F, Landos M, Bansemer MS, Stone DAJ, Qin JG, Oxley APA (2018) The inner workings of the outer surface: skin and gill microbiota as indicators of changing gut health in yellowtail kingfish. Front. Microbiol. 8:2664. https://doi.org/10.3389/fmicb.2017.02664
Marshall WS, Bellamy D (2010) The 50 year evolution of in vitro systems to reveal salt transport functions of teleost fish gills. Comp Biochem Physiol A Mol Integr Physiol 155:275–280. https://doi.org/10.1016/j.cbpa.2009.11.016
Peatman E, Lange M, Zhao H, Beck BH (2015) Physiology and immunology of mucosal barriers in catfish (Ictalurus spp.). Tissue barriers 3:e1068907. https://doi.org/10.1080/21688370.2015.1068907
Derome N, Gauthier J, Boutin S, Llewellyn M (2016) Bacterial opportunistic pathogens of fish. In: Hurst CJ (ed) The Rasputin effect: when commensals and symbionts become parasitic. Springer International Publishing, Cham, pp 81–108
Boutin S, Audet C, Derome N (2013) Probiotic treatment by indigenous bacteria decreases mortality without disturbing the natural microbiota of Salvelinus fontinalis. Canadian J Microbiol 59:662–670
Ye L, Amberg J, Chapman D, Gaikowski M, Liu WT (2014) Fish gut microbiota analysis differentiates physiology and behavior of invasive Asian carp and indigenous American fish. ISME J 8:541–551
Chiarello M, Villéger S, Bouvier C, Bettarel Y, Bouvier T (2015) High diversity of skin-associated bacterial communities of marine fishes is promoted by their high variability among body parts, individuals and species. FEMS Microbiol Ecol 91:fiv061
Sullam KE, Rubin BE, Dalton CM, Kilham SS, Flecker AS, Russell JA (2015) Divergence across diet, time and populations rules out parallel evolution in the gut microbiomes of Trinidadian guppies. ISME J 9:1508–1522. https://doi.org/10.1038/ismej.2014.1231
McGeoch MA, Chown SL (1998) Scaling up the value of bioindicators. Trend Ecol Evol 13:46–47
Legendre P, Legendre L (2012) Numerical ecology, 3rdEnglish edn. Elsevier Science BV, Amsterdam
Chiba SN, Iwatsuki Y, Yoshino T, Hanzawa N (2009) Comprehensive phylogeny of the family Sparidae (Perciformes: Teleostei) inferred from mitochondrial gene analyses. Genes Genet Syst 84:153–170
Louisi P (2015) Europe and Mediterranean marine fish identification guide. Eugen Ulmer, Paris
Pallaoro A, Dulcic S, Matic–Skoko M, Kraljevic M, Jardas I (2008) Biology of the salema Sarpa salpa (L. 1758) (Pisces, Sparidae) from the middle eastern Adriatic. J. Appl. Ichthyol. 24:276–281
Savage DC (1977) Microbial ecology of the gastrointestinal tract. Annu. Rev. Microbiol. 31:107–133
Vaahtovuo J, Toivanen P, Eerola E (2001) Study of murine fecal microflora by cellular fatty acids analysis; effect of age and mouse strain. A Van Leeuw 80:35–42
Parada AE, Needham DM, Fuhrman JA (2016) Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol. 18:1403–1414
Suzuki M, Giovannoni SJ (1996) Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl environ Microbiol 62:625
Edgar RC (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R (2010) QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7:335–336. https://doi.org/10.1038/nmeth.f.303
DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL (2006) Greengenes, a Chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Env Microbiol 72:5069–5072
R Core Team (2018) The R project for statistical computing. https://www.r-project.org
Oksanen J, Blanchet FG, Friendly M et al (2018) Vegan: community ecology package. R package version 2:5–2 https://CRAN.R-project.org/package=vegan
Sokal RR, Rohlf FJ (2015) Biometry. The principles and practice of statistics in biological research4th edn. W.H. Freeman and Company, New York
Statsoft Inc (2005) STATISTICA (data analysis software system), version 7.1
McMurdie PJ, Holmes S (2013) Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. Plos one 8:e61217
De Cáceres M, Legendre P (2009) Associations between species and groups of sites: indices and statistical inference. Ecology 90:3566–3574
De Cáceres M, Legendre P, Moretti M (2010) Improving indicator species analysis by combining groups of sites. Oikos 119:1674–1684
Dufrêne M, Legendre P (1997) Species assemblages and indicator species: the need for a flexible asymmetrical approach. Ecol. Monogr. 67:345–366
Crespo BG, Pommier T, Fernández-Gómez B, Pedrós-Alio P (2013) Taxonomic composition of the particle-attached and free-living bacterial assemblages in the Northwest Mediterranean Sea analyzed by pyrosequencing of the 16S rRNA. Microbiol Open 2:541–552
Semova I, Carten JD, Stombaugh J, Mackey LC, Knight R, Farber SA, Rawls JF (2012) Microbiota regulate intestinal absorption and metabolism of fatty acids in the zebrafish. Cell Host Microbe 12:277–288
Navarrete P, Magne F, Araneda C, Fuentes P, Barros L, Opazo R, Espejo R, Romero J (2012) PCR-TTGE analysis of 16S rRNA from rainbow trout (Oncorhynchus mykiss) gut microbiota reveals host-specific communities of active bacteria. PLoS One 7:e31335
Franchini P, Fruciano C, Frickey T, Jones JC, Meyer A (2014) The gut microbial community of Midas cichlid fish in repeatedly evolved limnetic–benthic species pairs. PLoS One 9:e95027
Larsen AM, Bullard SA, Womble M, Arias CR (2015) Community structure of skin microbiome of gulf killifish, Fundulus grandis, is driven by seasonality and not exposure to oiled sediments in a Louisiana salt marsh. Microb. Ecol. 70:534–544
Schmidt VT, Smith KF, Melvin DW, Amaral-Zettler LA (2015) Community assembly of a euryhaline fish microbiome during salinity acclimation. Mol. Ecol. 24:2537–2550
Smith CJ, Danilowicz BS, Meijer WG (2007) Characterization of the bacterial community associated with the surface and mucus layer of whiting (Merlangius merlangus). FEMS Microbiol. Ecol. 62:90–97
Larsen A, Tao Z, Bullard SA, Arias CR (2013) Diversity of the skin microbiota of fishes: evidence for host species specificity. FEMS Microbiol. Ecol. 85:483–494
Qin C, Huang K, Xu H (2002) Isolation and characterization of a novel polysaccharide from the mucus of the loach, Misgurnus anguillicaudatus. Carbohydr. Polym. 49:367–371
Zhang C, Huang KX (2005) Characteristic immunostimulation by MAP, a polysaccharide isolated from the mucus of the loach, Misgurnus anguillicaudatus. Carbohydr. Polym. 59:75–82
Lowrey L, Woodhams DC, Tacchi L, Salinas I (2015) Topographical mapping of the rainbow trout (Oncorhynchus mykiss) microbiome reveals a diverse bacterial community with antifungal properties in the skin. Appl Env Microbiol 81:6915–6925
Gómez GD, Balcázar JL (2018) A review on the interactions between gut microbiota and innate immunity of fish. FEMS Immunol. Med. Microbiol. 52:145–154 http://www.ncbi.nlm.nih.gov/pubmed/18081845
Gallet A, Koubbi P, Léger N, Scheifler M, Ruiz-Rodríguez M, Suzuki MT, Desdevises Y, Duperron S (2019) Low-diversity bacterial microbiota in Southern Ocean representatives of lanternfish genera Electrona, Protomyctophum and Gymnoscopelus (family Myctophidae). PLoS One 14(12):e0226159. https://doi.org/10.1371/journal.pone.0226159
Roeselers G, Mittge EK, Stephens WZ, Parichy DM, Cavanaugh CM, Guillemin K, Rawls JF (2011) Evidence for a core gut microbiota in the zebrafish. ISME J 5:1595–1608 http://www.ncbi.nlm.nih.gov/pubmed/21472014
Tremaroli V, Backhed F (2012) Functional interactions between the gut microbiota and host metabolism. Nature 489:242–249. https://doi.org/10.1038/nature11552
Acknowledgments
Fish sampling was performed by the “Service des Moyens à la Mer (FR3724) de l’Observatoire Océanologique de Banyuls/Mer (OOB)”. We are grateful to the BIO2MAR platform (http://bio2mar.obs-banyuls.fr) for providing technical help and support and access to instrumentation.
Funding
This work was funded by Sorbonne Université, programme Emergence 2016 (project SU-16-R-EMR-22-MICROFISH), which included a post-doctoral contract for MRR.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical Statement
All applicable international, national, and/or institutional guidelines for the use of animals were followed. The Observatoire Océanologique de Banyuls sur Mer holds the authorization from the “Direction interrégionale de la Mer Méditerrannée” for fishing and handling wild Mediterranean teleosts. Wild fish were caught (see above for details) by competent persons on the research vessel “Nereis II” and in accordance with the European Union Regulations concerning the protection and welfare of experimental animals (European directive 91/492/CCE).
Rights and permissions
About this article
Cite this article
Ruiz-Rodríguez, M., Scheifler, M., Sanchez-Brosseau, S. et al. Host Species and Body Site Explain the Variation in the Microbiota Associated to Wild Sympatric Mediterranean Teleost Fishes. Microb Ecol 80, 212–222 (2020). https://doi.org/10.1007/s00248-020-01484-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-020-01484-y