Abstract
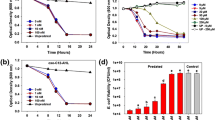
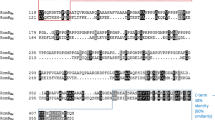
Bdellovibrio bacteriovorus is a predatory bacterium which lives by invading the periplasm of gram-negative bacteria and consuming them from within. This predator was thought to be dependent upon prey for nutrients since it lacks genes encoding for critical enzymes involved in amino acid biosynthesis. This study, however, found that planktonic attack-phase predators are not just dependent upon prey for nutrients, but rather, they respond to nutrients in the surrounding medium and, subsequently, synthesize and secrete proteases in a nutrient-dependent manner. The major secreted proteases were identified through mass spectrometry analyses. Subsequent RT-qPCR analyses found that the nutrient-induced proteases are similar to those expressed within the prey periplasm during the late intraperiplasmic growth phase. Furthermore, RNA sequencing found that incubating the planktonic attack-phase cells in a nutritious environment for a short period of time (4 h) changes its gene expression pattern to a status that is akin to the late intraperiplasmic phase, with more than 94% of the genes previously identified as being late intraperiplasmic-specific also being induced by nutrient broth in this study. This strong correlation between the gene expression patterns hints that the availability of hydrolyzed prey cell components to the predator is likely the stimulus controlling the expression of late intraperiplasmic B. bacteriovorus genes during predation.



Similar content being viewed by others
References
Stolp H, Starr MP (1963) Bdellovibrio bacteriovorus gen. et sp. n., a predatory, ectoparasitic, and bacteriolytic microorganism. Antonie van Leeuwenhoek 29:217–248
Dwidar M, Monnappa AK, Mitchell RJ (2012) The dual probiotic and antibiotic nature of Bdellovibrio bacteriovorus. BMB Rep. 45:71–78
Sockett RE (2009) Predatory lifestyle of Bdellovibrio bacteriovorus. Ann Rev Microbiol 63:523–539
Chen H, Brinkac LM, Mishra P, Li N, Lymperopoulou DS, Dickerson TL, Gordon-Bradley N, Williams HN, Badger JH (2015) Draft genome sequences for the obligate bacterial predators Bacteriovorax spp. of four phylogenetic clusters. Stand Genomic Sci 10:11
Pasternak Z, Njagi M, Shani Y, Chanyi R, Rotem O, Lurie-Weinberger MN, Koval S, Pietrokovski S, Gophna U, Jurkevitch E (2014) In and out: an analysis of epibiotic vs periplasmic bacterial predators. ISME J 8:625–635
Pasternak Z, Pietrokovski S, Rotem O, Gophna U, Lurie-Weinberger MN, Jurkevitch E (2013) By their genes ye shall know them: genomic signatures of predatory bacteria. ISME J 7:756–769
Rendulic S, Jagtap P, Rosinus A, Eppinger M, Baar C, Lanz C, Keller H, Lambert C, Evans KJ, Goesmann A, Meyer F, Sockett RE, Schuster SC (2004) A predator unmasked: life cycle of Bdellovibrio bacteriovorus from a genomic perspective. Science 303:689–692
Hobley L, Fung RKY, Lambert C, Harris MATS, Dabhi JM, King SS, Basford SM, Uchida K, Till R, Ahmad R, Aizawa S, Gomelsky M, Sockett RE (2012) Discrete cyclic di-GMP-dependent control of bacterial predation versus axenic growth in Bdellovibrio bacteriovorus. PLoS Pathog 8:e1002493
Karunker I, Rotem O, Dori-Bachash M, Jurkevitch E, Sorek R (2013) A global transcriptional switch between the attack and growth forms of Bdellovibrio bacteriovorus. PLoS One 8:e61850
Rotem O, Pasternak Z, Shimoni E, Belausov E, Porat Z, Pietrokovski S, Jurkevitch E (2015) Cell-cycle progress in obligate predatory bacteria is dependent upon sequential sensing of prey recognition and prey quality cues. P Natl Acad Sci USA 112:E6028–E6037
Lambert C, Evans KJ, Till R, Hobley L, Capeness M, Rendulic S, Schuster SC, Aizawa SI, Sockett RE (2006) Characterizing the flagellar filament and the role of motility in bacterial prey-penetration by Bdellovibrio bacteriovorus. Mol Microbiol 60:274–286
Lambert C, Chang CY, Capeness MJ, Sockett RE (2010) The first bite— profiling the predatosome in the bacterial pathogen Bdellovibrio. PLoS One 5:e8599
Monnappa AK, Dwidar M, Seo JK, Hur JH, Mitchell RJ (2014) Bdellovibrio bacteriovorus inhibits Staphylococcus aureus biofilm formation and invasion into human epithelial cells. Sci rep 4:3811
Spain EM, Nunez ME, Kim HJ, Taylor RJ, Thomas N, Wengen MB, Dalleska NF, Bromley JP, Schermerhorn KH, Ferguson MA (2016) Identification and differential production of ubiquinone-8 in the bacterial predator Bdellovibrio bacteriovorus. Res Microbiol 167:413–423
Seidler RJ, Starr MP (1969) Isolation and characterization of host-independent Bdellovibrios. J Bacteriol 100:769–785
Cotter TW, Thomashow MF (1992) Identification of a Bdellovibrio bacteriovorus genetic-locus, hit, associated with the host-independent phenotype. J Bacteriol 174:6018–6024
Im H, Kim D, Ghim CM, Mitchell RJ (2014) Shedding light on microbial predator-prey population dynamics using a quantitative bioluminescence assay. Microbial Ecol 67:167–176
Dwidar M, Leung BM, Yaguchi T, Takayama S, Mitchell RJ (2013) Patterning bacterial communities on epithelial cells. PLoS One 8:e67165
Dwidar M, Hong S, Cha M, Jang J, Mitchell RJ (2012) Combined application of bacterial predation and carbon dioxide aerosols to effectively remove biofilms. Biofouling 28:671–680
Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M (2006) In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protocols 1:2856–2860
Nesvizhskii AI, Keller A, Kolker E, Aebersold R (2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem 75:4646–4658
Untergasser A (2008) RNAprep-trizol combined with columns. http://www.untergasser.de/lab/protocols/rna_prep_comb_trizol_v1_0.htm.
Monnappa AK, Dwidar M, Mitchell RJ (2013) Application of bacterial predation to mitigate recombinant bacterial populations and their DNA. Soil Biol Biochem 57:427–435
Dwidar M, Nam D, Mitchell RJ (2014) Indole negatively impacts predation by Bdellovibrio bacteriovorus and its release from the bdelloplast. Environ Microbiol 4:1009–1022
Mortazavi A, Williams BA, Mccue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5:621–628
Anders S, Huber W (2010) Differential expression analysis for sequence count data. Genome Biol 11:R106
Robinson MD, Oshlack A (2010) A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol 11:R25
Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B (2000) Artemis: sequence visualization and annotation. Bioinformatics 16:944–945
Fenton AK, Lambert C, Wagstaff PC, Sockett RE (2010) Manipulating each MreB of Bdellovibrio bacteriovorus gives diverse morphological and predatory phenotypes. J Bacteriol 192:1299–1311
Roschanski N, Klages S, Reinhardt R, Linscheid M, Strauch E (2011) Identification of genes essential for prey-independent growth of Bdellovibrio bacteriovorus HD100. J Bacteriol 193:1745–1756
Capeness MJ, Lambert C, Lovering AL, Till R, Uchida K, Chaudhuri R, Alderwick LJ, Lee DJ, Swarbreck D, Liddell S, Aizawa SI, Sockett RE (2013) Activity of Bdellovibrio hit locus proteins, Bd0108 and Bd0109, links type IVa pilus extrusion/retraction status to prey-independent growth signalling. PLoS One 8:e79759
Schwudke D, Bernhardt A, Beck S, Madela K, Linscheid MW, Appel B, Strauch E (2005) Transcriptional activity of the host-interaction locus and a putative pilin gene of Bdellovibrio bacteriovorus in the predatory life cycle. Curr Microbiol 51:310–316
Nunez ME, Martin MO, Chan PH, Spain EM (2005) Predation, death, and survival in a biofilm: Bdellovibrio investigated by atomic force microscopy. Colloid Surface B 42:263–271
Ferguson MA, Nunez ME, Kim HJ, Goffredi S, Shamskhou E, Faudree L, Chang E, Landry RM, Ma A, Choi DE, Thomas N, Schmitt J, Spain EM (2014) Spatially organized films from Bdellovibrio bacteriovorus prey lysates. Appl Environ Microb 80:7405–7414
Amy PS, Morita RY (1983) Starvation-survival patterns of sixteen freshly isolated open-ocean bacteria. Appl Environ Microbiol 45:1109–1115
Hood MA, Macdonell MT (1987) Distribution of ultramicrobacteria in a gulf coast estuary and induction of ultramicrobacteria. Microb Ecol 14:113–127
Reeve CA, Bockman AT, Matin A (1984) Role of protein degradation in the survival of carbon-starved Escherichia coli and Salmonella typhimurium. J Bacteriol 157:758–763
Rybkin AI, Ravin VK (1987) Decreased synthetic activity as a possible cause of the death of Escherichia coli bacteria during amino acid starvation. Mikrobiologiia 56:227–231
Postgate JR, Hunter JR (1962) The survival of starved bacteria. J Gen Microbiol 29:233–263
Gray KM, Ruby EG (1990) Prey-derived signals regulating duration of the developmental growth-phase of Bdellovibrio bacteriovorus. J Bacteriol 172:4002–4007
Acknowledgements
This work was generously supported by the Korea Health Industry Development Institute (KHIDI) for financial support (Grant No. HI13C13550000). We wish to thank the UNIST Central Research Facilities (UCRF) for their help with the LC-MS/MS data.
Author information
Authors and Affiliations
Contributions
M.D. and H.I. designed and carried out the experiments. R.J.M. supervised the experimental work. M.D., H.I., and R.J.M. evaluated the data. M.D. and J.S.K. performed the mass spectrometry experiments. J.S.K. performed the mass spectrometry analyses. M.D., H.I., J.S.K., and R.J.M. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Rights and permissions
About this article
Cite this article
Dwidar, M., Im, H., Seo, J.K. et al. Attack-Phase Bdellovibrio bacteriovorus Responses to Extracellular Nutrients Are Analogous to Those Seen During Late Intraperiplasmic Growth. Microb Ecol 74, 937–946 (2017). https://doi.org/10.1007/s00248-017-1003-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-017-1003-1