Abstract
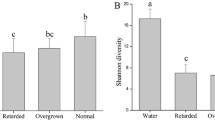
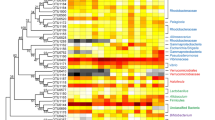
Increasing evidence of tight links among the gut microbiota, obesity, and host health has emerged, but knowledge of the ecological processes that shape the variation in microbial assemblages across growth rates remains elusive. Moreover, inadequately control for differences in factors that profoundly affect the gut microbial community, hampers evaluation of the gut microbiota roles in regulating growth rates. To address this gap, we evaluated the composition and ecological processes of the gut bacterial community in cohabitating retarded, overgrown, and normal shrimps from identically managed ponds. Gut bacterial community structures were distinct (P = 0.0006) among the shrimp categories. Using a structural equation modeling (SEM), we found that changes in the gut bacterial community were positively related to digestive activities, which subsequently affected shrimp growth rate. This association was further supported by intensified interspecies interaction and enriched lineages with high nutrient intake efficiencies in overgrown shrimps. However, the less phylogenetic clustering of gut microbiota in overgrown and retarded subjects may offer empty niches for pathogens invasion, as evidenced by higher abundances of predicted functional pathways involved in disease infection. Given no differences in biotic and abiotic factors among the cohabitating shrimps, we speculated that the distinct gut community assembly could be attributed to random colonization in larval shrimp (e.g., priority effects) and that an altered microbiota could be a causative factor in overgrowth or retardation in shrimp. To our knowledge, this is the first study to provide an integrated overview of the direct roles of gut microbiota in shaping shrimp growth rate and the underlying ecological mechanisms.






Similar content being viewed by others
References
Sommer F, Bäckhed F (2013) The gut microbiota—masters of host development and physiology. Nat Rev Microbiol 11:227–238
Tap J, Furet JP, Bensaada M, Philippe C, Roth H, Rabot S, Lakhdari O, Lombard V, Henrissat B, Corthier G (2015) Gut microbiota richness promotes its stability upon increased dietary fibre intake in healthy adults. Environ. Microbiol. 17:4954–4964
Xiong J, Wang K, Wu J, Qiuqian L, Yang K, Qian Y, Zhang D (2015) Changes in intestinal bacterial communities are closely associated with shrimp disease severity. Appl. Microbiol. Biotechnol. 99:6911–6919
Mosca A, Leclerc M, Hugot JP (2016) Gut microbiota diversity and human diseases: should we reintroduce key predators in our ecosystem? Front. Microbiol. 7:455
Yan Q, Li J, Yu Y, Wang J, He Z, Van Nostrand JD, Kempher ML, Wu L, Wang Y, Liao L, Li X, Wu S, Ni J, Wang C, Zhou J (2016) Environmental filtering decreases with fish development for the assembly of gut microbiota. Environ. Microbiol. doi:10.1111/1462-2920.13365
Ng'ambi J, Li R, Mu C, Song W, Liu L, Wang C (2016) Dietary administration of saponin stimulates growth of the swimming crab Portunus Trituberculatus and enhances its resistance against vibrio alginolyticus infection. Fish Shellfish Immunol 59:1050–4648
Xiong J, Zhu J, Zhang D (2014) The application of bacterial indicator phylotypes to predict shrimp health status. Appl. Microbiol. Biotechnol. 98:8291–8299
Sabes-Figuera R, Knapp M, Bendeck M, Mompart-Penina A, Salvador-Carulla L (2015) The composition of the gut microbiota throughout life, with an emphasis on early life. Microb. Ecol. Health Dis. 26:24–29
Burns AR, Stephens WZ, Stagaman K, Wong S, Rawls JF, Guillemin K, Bohannan BJM (2016) Contribution of neutral processes to the assembly of gut microbial communities in the zebrafish over host development. ISME J 10:655–664
Ramayo-Caldas Y, Mach N, Lepage P, Levenez F, Denis C, Lemonnier G, Leplat JJ, Billon Y, Berri M, Doré J, Rogel-Gaillard C, Estellé J (2016) Phylogenetic network analysis applied to pig gut microbiota identifies an ecosystem structure linked with growth traits. ISME J 10:2973–2977
Miyanaga A, Shimizu K, Noro R, Seike M, Kitamura K, Kosaihira S, Minegishi Y, Shukuya T, Yoshimura A, Kawamoto M (2011) Enterotypes of the human gut microbiome. Nature 473:174–180
Sha Y, Liu M, Wang B, Jiang K, Sun G, Wang L (2016) Gut bacterial diversity of farmed sea cucumbers Apostichopus japonicus with different growth rates. Microbiology 85:109–115
Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI (2006) An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444:1027–1031
Riva A, Borgo F, Lassandro C, Verduci E, Morace G, Borghi E, Berry D (2016) Pediatric obesity is associated with an altered gut microbiota and discordant shifts in Firmicutes populations. Environ. Microbiol. doi:10.1111/1462-2920.13463
Holmes E, Li JV, Marchesi JR, Nicholson JK (2012) Gut microbiota composition and activity in relation to host metabolic phenotype and disease risk. Cell Metab. 16:559–564
Ferrer M, Ruiz A, Lanza F, Haange SB, Oberbach A, Till H, Bargiela R, Campoy C, Segura MT, Richter M (2013) Microbiota from the distal guts of lean and obese adolescents exhibit partial functional redundancy besides clear differences in community structure. Environ. Microbiol. 15:211–226
Sullam KE, Essinger SD, Lozupone CA, O’Connor MP, Rosen GL, Knight R, Kilham SS, Russell JA (2012) Environmental and ecological factors that shape the gut bacterial communities of fish: a meta-analysis. Mol. Ecol. 21:3363–3378
Wiens JJ, Ackerly DD, Allen AP, Anacker BL, Buckley LB, Cornell HV, Damschen EI, Davies TJ, Grytnes JA, Harrison SP (2010) Niche conservatism as an emerging principle in ecology and conservation biology. Ecol. Lett. 13:1310–1324
Zhu J, Dai W, Qiu Q, Dong C, Zhang J, Xiong J (2016) Contrasting ecological processes and functional compositions between intestinal bacterial community in healthy and diseased shrimp. Microb. Ecol. 72:975–985
Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, Egholm M, Henrissat B, Heath AC, Knight R, Gordon JI (2009) A core gut microbiome in obese and lean twins. Nature 457:480–484
Duran-Pinedo AE, Paster B, Teles R, Friaslopez J (2011) Correlation network analysis applied to complex biofilm communities. PLoS One 6:e28438
Hallam SJ, McCutcheon JP (2015) Microbes don’t play solitaire: how cooperation trumps isolation in the microbial world. Environ. Microbiol. Rep. 7:26–28
Egan S, Gardiner M (2016) Microbial dysbiosis: rethinking disease in marine ecosystems. Front. Microbiol. 7:991
Faust K, Raes J (2012) Microbial interactions: from networks to models. Nat Rev Microbiol 10:538–550
Xiong J, Chen H, Hu C, Ye X, Kong D, Zhang D (2015) Evidence of bacterioplankton community adaptation in response to long-term mariculture disturbance. Sci Rep 5:15274
Herlemann DP, Labrenz M, Jürgens K, Bertilsson S, Waniek JJ, Andersson AF (2011) Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J 5:1571–1579
Magoč T, Salzberg SL (2011) FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27:2957–2963
Caporaso JG, Kuczynski J, Stombaugh J (2010) QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7:335–336
Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200
Edgar RC (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461
De Santis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL (2006) Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Applied Environ Microbiol 72:5069–5072
De Santis TZ, Hugenholtz P, Keller K, Brodie EL, Larsen N, Piceno YM, Phan R, Andersen GL (2006) NAST: a multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res. 34:394–399
Price MN, Dehal PS, Arkin AP (2009) FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 26:1641–1650
Clarke KR (1993) Non-parametric multivariate analyses of changes in community structure. Aust. J. Ecol. 18:117–143
R Development Core Team (2013) R: A language and environment for statistical computing
Dufrêne M, Legendre P (1997) Species assemblages and indicator species: the need for a flexible asymmetrical approach. Ecol. Monogr. 67:345–366
Roberts DW (2007) labdsv: Ordination and multivariate analysis for ecology. R package version
Chase JM, Kraft NJ, Smith KG, Vellend M, Inouye BD (2011) Using null models to disentangle variation in community dissimilarity from variation in α-diversity. Ecosphere 2:art24
Stegen JC, Lin X, Fredrickson JK, Chen X, Kennedy DW, Murray CJ, Rockhold ML, Konopka A (2013) Quantifying community assembly processes and identifying features that impose them. ISME J 7:2069–2079
Kembel SW, Cowan PD, Helmus MR, Cornwell WK, Morlon H, Ackerly DD, Blomberg SP, Webb CO (2010) Picante: R tools for integrating phylogenies and ecology. Bioinformatics 26:1463–1464
Webb CO, Ackerly DD, McPeek MA, Donoghue MJ (2002) Phylogenies and community ecology. Ann Rev Ecol Syst 33:475–505
Deng Y, Jiang Y, Yang Y, He Z, Luo F, Zhou J (2012) Molecular ecological network analyses. BMC Bioinformatics 13:113
Newman MEJ (2003) The structure and function of complex networks. SIAM Rev. 45:167–256
Shannon P, Markiel A, Ozier O, Baliga NS, Wang J, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13:2498–2504
Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Thurber RLV, Knight R (2013) Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotech 31:814–821
Wu J, Xiong J, Hu C, Shi Y, Wang K, Zhang D (2015) Temperature sensitivity of soil bacterial community along contrasting warming gradient. Appl. Soil Ecol. 94:40–48
Newman ME (2006) Modularity and community structure in networks. Proc Nat Acad Sci USA 103:8577–8582
Guimera R, Sales-Pardo M, Amaral LA (2007) Classes of complex networks defined by role-to-role connectivity profiles. Nat Physcis 3:63–69
Bäckhed F, Fraser C, Ringel Y, Sanders ME, Sartor RB, Sherman P, Versalovic J, Young V, Finlay BB (2012) Defining a healthy human gut microbiome: current concepts, future directions, and clinical applications. Cell Host Microbe 12:611–622
Gilbert JA, Quinn RA, Debelius J, Xu ZZ, Morton J, Garg N, Jansson JK, Dorrestein PC, Knight R (2016) Microbiome-wide association studies link dynamic microbial consortia to disease. Nature 535:94–103
Xiong J, Dai W, Li C (2016) Advances, challenges, and directions in shrimp disease control: the guidelines from an ecological perspective. Appl Microbiol Biotech 100:6947–6954
Fukami T (2015) Historical contingency in community assembly: integrating niches, species pools, and priority fffects. Ann Rev Ecol Evol Syst 46:1–23
Fjellheim AJ, Playfoot KJ, Skjermo J, Vadstein O (2012) Inter-individual variation in the dominant intestinal microbiota of reared Atlantic cod (Gadus morhua L.) larvae. Aquac. Res. 43:1499–1508
Harakeh SM, Khan I, Kumosani T, Barbour E, Almasaudi SB, Bahijri SM, Alfadul SM, Ajabnoor GMA, Azhar EI (2016) Gut microbiota: a contributing factor to obesity. Front Cell Infet Microbiol 6:95
Mcilroy SJ, Nielsen PH (2014) The family Saprospiraceae. pp 863–889
Galitskaya P, Akhmetzyanova L, Selivanovskaya S (2016) Biochar carrying hydrocarbon decomposers promotes degradation during the early stage of bioremediation. Biogeosci Discuss 13:5739–5752
Lebba V, Santangelo F, Totino V, Nicoletti M, Gagliardi A, De Biase RV, Cucchiara S, Nencioni L, Conte MP, Schippa S (2013) Higher prevalence and abundance of Bdellovibrio bacteriovorus in the human gut of healthy subjects. PLoS One 8:e61608
Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R (2012) Diversity, stability and resilience of the human gut microbiota. Nature 489:220–230
Stecher B, Chaffron S, Käppeli R, Hapfelmeier S, Freedrich S, Weber TC, Kirundi J, Suar M, Mccoy KD, Von MC (2010) Like will to like: abundances of closely related species can predict susceptibility to intestinal colonization by pathogenic and commensal bacteria. PLoS Path 6:e1000711
Wagner A (2011) Genotype networks shed light on evolutionary constraints. Trends Ecol. Evol. 26:577–584
Adler PB, Hillerislambers J, Levine JM (2007) A niche for neutrality. Ecol. Lett. 10:95–104
Mayfield MM, Levine JM (2010) Opposing effects of competitive exclusion on the phylogenetic structure of communities. Ecol. Lett. 13:1085–1093
Inglis RF, Gardner A, Cornelis P, Buckling A (2009) Spite and virulence in the bacterium Pseudomonas Aeruginosa. Proc Nat Acad Sci USA 106:5703–5707
Jousset A, Schulz W, Scheu S, Eisenhauer N (2011) Intraspecific genotypic richness and relatedness predict the invasibility of microbial communities. ISME J 5:1108–1114
Yiu JHC, Dorweiler B, Woo CW (2016) Interaction between gut microbiota and toll-like receptor: from immunity to metabolism. J Mol Med. doi:10.1007/s00109-016-1474-4
Mallon CA, Elsas JDV, Salles JF (2015) Microbial invasions: the process, patterns, and mechanisms. Trends Microbiol. 23:719–729
Acknowledgements
This work was supported by the Zhejiang Province Public Welfare Technology Application Research Project (2016C32063), and the Open Fund of Key Laboratory of Marine Biogenetic Resources, Third Institute of Oceanography (HY201601); the Project of Science and Technology Department of Ningbo (2015C10062); and the KC Wong Magna Fund in Ningbo University.
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
ESM 1
(PDF 284 kb)
Rights and permissions
About this article
Cite this article
Xiong, J., Dai, W., Zhu, J. et al. The Underlying Ecological Processes of Gut Microbiota Among Cohabitating Retarded, Overgrown and Normal Shrimp. Microb Ecol 73, 988–999 (2017). https://doi.org/10.1007/s00248-016-0910-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-016-0910-x