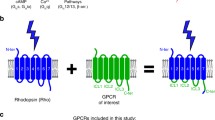
Abstract
Eukaryotic cells use G protein-coupled receptors (GPCRs) to convert external stimuli into internal signals to elicit cellular responses. However, how mutations in GPCR-coding genes affect GPCR activation and downstream signaling pathways remain poorly understood. Approaches such as deep mutational scanning show promise in investigations of GPCRs, but a high-throughput method to measure rhodopsin activation has yet to be achieved. Here, we scale up a fluorescent reporter assay in budding yeast that we engineered to study rhodopsin’s light-activated signal transduction. Using this approach, we measured the mutational effects of over 1200 individual human rhodopsin mutants, generated by low-frequency random mutagenesis of the GPCR rhodopsin (RHO) gene. Analysis of the data in the context of rhodopsin’s three-dimensional structure reveals that transmembrane helices are generally less tolerant to mutations compared to flanking helices that face the lipid bilayer, which suggest that mutational tolerance is contingent on both the local environment surrounding specific residues and the specific position of these residues in the protein structure. Comparison of functional scores from our screen to clinically identified rhodopsin disease variants found many pathogenic mutants to be loss of function. Lastly, functional scores from our assay were consistent with a complex counterion mechanism involved in ligand-binding and rhodopsin activation. Our results demonstrate that deep mutational scanning is possible for rhodopsin activation and can be an effective method for revealing properties of mutational tolerance that may be generalizable to other transmembrane proteins.





Similar content being viewed by others
References
Adkar BV, Tripathi A, Sahoo A et al (2012) Protein model discrimination using mutational sensitivity derived from deep sequencing. Structure 20:371–381. https://doi.org/10.1016/j.str.2011.11.021
Antonarakis SE, Beckmann JS (2006) Focus on monogenic disorders. Nat Rev Genet 7:277–282. https://doi.org/10.1038/nrg1826
Athanasiou D, Aguila M, Bellingham J et al (2018) The molecular and cellular basis of rhodopsin retinitis pigmentosa reveals potential strategies for therapy. Prog Retin Eye Res 62:1–23. https://doi.org/10.1016/j.preteyeres.2017.10.002
Bendell CJ, Liu S, Aumentado-Armstrong T et al (2014) Transient protein-protein interface prediction: datasets, features, algorithms, and the RAD-T predictor. BMC Bioinform 15:1–12. https://doi.org/10.1186/1471-2105-15-82
Bowie JU, Reidhaar-Olson JF, Lim WA, Sauer RT (1990) Deciphering the message in protein sequences: tolerance to amino acid substitutions. Science 247:1306–1310. https://doi.org/10.1126/science.2315699
Chen Y, Jastrzebska B, Cao P et al (2014) Inherent instability of the retinitis pigmentosa P23H mutant opsin. J Biol Chem 289:9288–9303. https://doi.org/10.1074/jbc.M114.551713
Chothia C (1975) Structural invariants in protein folding. Nature 254:304–308. https://doi.org/10.1038/254304a0
Cohen GB, Oprian DD, Robinson PR (1992) Mechanism of activation and inactivation of Opsin: role of Glu113 and Lys296. Biochemistry 31:12592–12601. https://doi.org/10.1021/bi00165a008
Dong S, Rogan SC, Roth BL (2010) Directed molecular evolution of DREADDs: a generic approach to creating next-generation RASSLs. Nat Protoc 5:561–573. https://doi.org/10.1038/nprot.2009.239
Doolan KM, Colby DW (2015) Conformation-dependent epitopes recognized by prion protein antibodies probed using mutational scanning and deep sequencing. J Mol Biol 427:328–340. https://doi.org/10.1016/j.jmb.2014.10.024
Duncan AL, Song W, Sansom MSP (2019) Lipid-dependent regulation of ion channels and G protein-coupled receptors: insights from structures and simulations. Annu Rev Pharmacol Toxicol 60:31–50. https://doi.org/10.1146/annurev-pharmtox-010919
Eilers M, Shekar SC, Shieh T et al (2000) Internal packing of helical membrane proteins. Proc Natl Acad Sci 97:5796–5801. https://doi.org/10.1073/pnas.97.11.5796
Elazar A, Weinstein J, Biran I et al (2016) Mutational scanning reveals the determinants of protein insertion and association energetics in the plasma membrane. Elife 5:1–19. https://doi.org/10.7554/elife.12125
Engelman DM, Zaccai G (1980) Bacteriorhodopsin is an inside-out protein. Proc Natl Acad Sci USA 77:5894–5898. https://doi.org/10.1073/pnas.77.10.5894
Erdogmus S, Storch U, Danner L, et al (2019) Helix 8 is the essential structural motif of mechanosensitive GPCRs. Nat Commun. https://doi.org/10.1038/s41467-019-13722-0
Ernst A, Gfeller D, Kan Z et al (2010) Coevolution of PDZ domain-ligand interactions analyzed by high-throughput phage display and deep sequencing. Mol Biosyst 6:1782–1790. https://doi.org/10.1039/c0mb00061b
Fokkema IFAC, Taschner PEM, Schaafsma GCP et al (2011) LOVD vol 2.0: The next generation in gene variant databases. Hum Mutat 32:557–563. https://doi.org/10.1002/humu.21438
Fowler DM, Araya CL, Fleishman SJ et al (2010) High-resolution mapping of protein sequence-function relationships. Nat Methods 7:741–746. https://doi.org/10.1038/nmeth.1492
Fowler DM, Stephany JJ, Fields S (2014) Measuring the activity of protein variants on a large scale using deep mutational scanning. Nat Protoc 9:2267–2284. https://doi.org/10.1038/nprot.2014.153
Gimpelev M, Forrest LR, Murray D, Honig B (2004) Helical packing patterns in membrane and soluble proteins. Biophys J 87:4075–4086. https://doi.org/10.1529/biophysj.104.049288
Glazer AM, Kroncke BM, Matreyek KA, et al (2020) Deep mutational scan of an SCN5A voltage sensor. Circ Genom Precis Med 20–29. https://doi.org/10.1161/CIRCGEN.119.002786
Gray VE, Hause RJ, Fowler DM (2017) Analysis of large-scale mutagenesis data to assess the impact of single amino acid substitutions. Genetics 207:53–61. https://doi.org/10.1534/genetics.117.300064
Guo HH, Choe J, Loeb LA (2004) Protein tolerance to random amino acid change. Proc Natl Acad Sci 101:9205–9210. https://doi.org/10.1073/pnas.0403255101
Hamosh A, Scott AF, Amberger JS et al (2005) Online mendelian inheritance in man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res 33:514–517. https://doi.org/10.1093/nar/gki033
Hargrave PA, McDowell JH, Curtis DR et al (1983) The structure of bovine rhodopsin. Biophys Struct Mech 9:235–244. https://doi.org/10.1007/BF00535659
Heydenreich FM, Vuckovic Z, Matkovic M, Veprintsev DB (2015) Stabilization of G protein-coupled receptors by point mutations. Front Pharmacol 6:1–15. https://doi.org/10.3389/fphar.2015.00082
Hietpas RT, Bank C, Jensen JD, Bolon DNA (2013) Shifting fitness landscapes in response to altered environments. Evolution (n y) 67:3512–3522. https://doi.org/10.1111/evo.12207
Janz JM, Farrens DL (2004) Role of the retinal hydrogen bond network in rhodopsin Schiff base stability and hydrolysis. J Biol Chem 279:55886–55894. https://doi.org/10.1074/jbc.M408766200
Jastrzebska B, Chen Y, Orban T et al (2015) Disruption of rhodopsin dimerization with synthetic peptides targeting an interaction interface. J Biol Chem 290:25728–25744. https://doi.org/10.1074/jbc.M115.662684
Jones EM, Lubock NB, Venkatakrishnan A et al (2020) Structural and functional characterization of G protein–coupled receptors with deep mutational scanning. Elife 9:1–28. https://doi.org/10.7554/eLife.54895
Kang Y, Kuybeda O, De Waal PW et al (2018) Cryo-EM structure of human rhodopsin bound to an inhibitory G protein. Nature 558:553–558. https://doi.org/10.1038/s41586-018-0215-y
Karczewski KJ, Francioli LC, Tiao G et al (2020) The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581:434–443. https://doi.org/10.1038/s41586-020-2308-7
Keen TJ, Inglehearn CF, Lester DH et al (1991) Autosomal dominant retinitis pigmentosa: four new mutations in rhodopsin, one of them in the retinal attachment site. Genomics 11:199–205. https://doi.org/10.1016/0888-7543(91)90119-Y
Kim JM, Altenbach C, Kono M et al (2004) Structural origins of constitutive activation in rhodopsin: role of the K296/E113 salt bridge. Proc Natl Acad Sci U S A 101:12508–12513. https://doi.org/10.1073/pnas.0404519101
Knepp AM, Periole X, Marrink SJ et al (2012) Rhodopsin forms a dimer with cytoplasmic helix 8 contacts in native membranes. Biochemistry 51:1819–1821. https://doi.org/10.1021/bi3001598
Kozek KA, Glazer AM, Ng CA et al (2020) High-throughput discovery of trafficking-deficient variants in the cardiac potassium channel KV11.1. Heart Rhythm 17:2180–2189. https://doi.org/10.1016/j.hrthm.2020.05.041
Krebs MP, Holden DC, Joshi P et al (2010) Molecular mechanisms of rhodopsin retinitis pigmentosa and the efficacy of pharmacological rescue. J Mol Biol 395:1063–1078. https://doi.org/10.1016/j.jmb.2009.11.015
Kuntz CP, Woods H, McKee AG et al (2022) Towards generalizable predictions for G protein-coupled receptor variant expression. Biophys J 121:2712–2720. https://doi.org/10.1016/J.BPJ.2022.06.018
Landrum MJ, Lee JM, Benson M et al (2018) ClinVar: Improving access to variant interpretations and supporting evidence. Nucleic Acids Res 46:D1062–D1067. https://doi.org/10.1093/nar/gkx1153
Leung NY, Montell C (2017) Unconventional Roles of Opsins. Annu Rev Cell Dev Biol 33:241–264. https://doi.org/10.1146/annurev-cellbio-100616-060432
Liang Y, Fotiadis D, Filipek S et al (2003) Organization of the G protein-coupled receptors rhodopsin and opsin in native membranes. J Biol Chem 278:21655–21662. https://doi.org/10.1074/jbc.M302536200
Lim WA, Sauer RT (1989) Alternative packing arrangements in the hydrophobic core of λrepresser. Nature 339:31–36. https://doi.org/10.1038/339031a0
Lim WA, Sauer RT (1991) The role of internal packing interactions in determining the structure and stability of a protein. J Mol Biol 219:359–376. https://doi.org/10.1016/0022-2836(91)90570-V
Lin Y-C, Guo YR, Miyagi A et al (2019) Force-induced conformational changes in PIEZO1. Nature 573:230–234. https://doi.org/10.1038/s41586-019-1499-2
Mckee AG, Kuntz CP, Ortega JT et al (2021) Systematic profiling of temperature- and retinal-sensitive rhodopsin variants by deep mutational scanning. J Biol Chem 297:101359. https://doi.org/10.1016/j.jbc.2021.101359
McLaughlin RN, Poelwijk FJ, Raman A et al (2012) The spatial architecture of protein function and adaptation. Nature 491:138–142. https://doi.org/10.1038/nature11500
Mollaaghababa R, Davidson FF, Kaiser C, Khorana HG (1996) Structure and function in rhodopsin: expression of functional mammalian opsin in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 93:11482–11486. https://doi.org/10.1073/pnas.93.21.11482
Nakayama TA, Khorana HG (1991) Mapping of the amino acids in membrane-embedded helices that interact with the retinal chromophore in bovine rhodopsin. J Biol Chem 266:4269–4275. https://doi.org/10.1016/S0021-9258(20)64317-4
Penn WD, McKee AG, Kuntz CP et al (2020) Probing biophysical sequence constraints within the transmembrane domains of rhodopsin by deep mutational scanning. Sci Adv 6:1–10. https://doi.org/10.1126/sciadv.aay7505
Pettersen EF, Goddard TD, Huang CC et al (2004) UCSF Chimera - a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. https://doi.org/10.1002/jcc.20084
Ramon E, Cordomi A, Aguilà M et al (2014) Differential light-induced responses in sectorial inherited retinal degeneration. J Biol Chem 289:35918–35928. https://doi.org/10.1074/jbc.M114.609958
Richards FM (1977) Areas, volumes, packing and protein structure. Annu Rev Biophys Bioeng 6:151–176
Richards S, Aziz N, Bale S et al (2015) Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the american college of medical genetics and genomics and the association for molecular pathology. Genet Med 17:405–424. https://doi.org/10.1038/gim.2015.30
Di Roberto RB, Scott BM, Peisajovich SG (2017) Directed Evolution Methods to Rewire Signaling Networks. 1596: 321–337
Robinson PR, Cohen GB, Zhukovsky EA, Oprian DD (1992) Constitutively active mutants of rhodopsin. Neuron 9:719–725. https://doi.org/10.1016/0896-6273(92)90034-B
Romero PA, Tran TM, Abate AR (2015) Dissecting enzyme function with microfluidic-based deep mutational scanning. Proc Natl Acad Sci U S A 112:7159–7164. https://doi.org/10.1073/pnas.1422285112
Roushar FJ, Mckee AG, Kuntz CP, et al (2022) Molecular basis for variations in the sensitivity of pathogenic rhodopsin variants to 9-cis-retinal. https://doi.org/10.1016/j.jbc.2022.102266
Sakami S, Kolesnikov AV, Kefalov VJ, Palczewski K (2014) P23H opsin knock-in mice reveal a novel step in retinal rod disc morphogenesis. Hum Mol Genet 23:1723–1741. https://doi.org/10.1093/hmg/ddt561
Sakmar TP, Franke RR, Khorana HG (1989) Glutamic acid-113 serves as the retinylidene Schiff base counterion in bovine rhodopsin. Proc Natl Acad Sci U S A 86:8309–8313. https://doi.org/10.1073/pnas.86.21.8309
Schlinkmann KM, Honegger A, Tureci E et al (2012) Critical features for biosynthesis, stability, and functionality of a G protein-coupled receptor uncovered by all-versus-all mutations. Proc Natl Acad Sci 109:9810–9815. https://doi.org/10.1073/pnas.1202107109
Scott BM, Chen SK, Bhattacharyya N et al (2019) Coupling of human rhodopsin to a yeast signaling pathway enables characterization of mutations associated with retinal disease. Genetics 211:597–615. https://doi.org/10.1534/genetics.118.301733
Shin HS, Cho Y, Choe DH et al (2014) Exploring the functional residues in a flavin-binding fluorescent protein using deep mutational scanning. PLOS ONE 9:1–10. https://doi.org/10.1371/journal.pone.0097817
Starita LM, Ahituv N, Dunham MJ et al (2017) Variant interpretation: functional assays to the rescue. Am J Hum Genet 101:315–325. https://doi.org/10.1016/j.ajhg.2017.07.014
Stoy H, Gurevich VV (2015) How genetic errors in GPCRs affect their function: possible therapeutic strategies. Genes Dis 2:108–132. https://doi.org/10.1016/j.gendis.2015.02.005
Sun S, Yang F, Tan G et al (2016) An extended set of yeast-based functional assays accurately identifies human disease mutations. Genome Res 26:670–680. https://doi.org/10.1101/gr.192526.115
Tam BM, Moritz OL (2006) Characterization of rhodopsin P23H-induced retinal degeneration in a Xenopus laevis model of retinitis pigmentosa. Invest Ophthalmol vis Sci 47:3234–3241. https://doi.org/10.1167/iovs.06-0213
Terakita A, Yamashita T, Shichida Y (2000) Highly conserved glutamic acid in the extracellular IV-V loop in rhodopsins acts as the counterion in retinochrome, a member of the rhodopsin family. Proc Natl Acad Sci 97:14263–14267. https://doi.org/10.1073/pnas.260349597
Terakita A, Koyanagi M, Tsukamoto H et al (2004) Counterion displacement in the molecular evolution of the rhodopsin family. Nat Struct Mol Biol 11:284–289. https://doi.org/10.1038/nsmb731
Tsai CJ, Marino J, Adaixo R et al (2019) Cryo-EM structure of the rhodopsin-Gαi- βγ complex reveals binding of the rhodopsin C-terminal tail to the gβ subunit. Elife 8:1–19. https://doi.org/10.7554/eLife.46041.001
Wan A, Place E, Pierce EA, Comander J (2019) Characterizing variants of unknown significance in rhodopsin: a functional genomics approach. Hum Mutat 40:1127–1144. https://doi.org/10.1002/humu.23762
Weile J, Roth FP (2018) Multiplexed assays of variant effects contribute to a growing genotype–phenotype atlas. Hum Genet 137:665–678. https://doi.org/10.1007/s00439-018-1916-x
Weile J, Sun S, Cote AG et al (2017) A framework for exhaustively mapping functional missense variants. Mol Syst Biol 13:957. https://doi.org/10.15252/msb.20177908
Yan ECY, Kazmi MA, Ganim Z et al (2003) Retinal counterion switch in the photoactivation of the G protein-coupled receptor rhodopsin. Proc Natl Acad Sci 100:9262–9267. https://doi.org/10.1073/pnas.1531970100
Yang T, Snider BB, Oprian DD (1997) Synthesis and characterization of a novel retinylamine analog inhibitor of constitutively active rhodopsin mutants found in patients with autosomal dominant retinitis pigmentosa. Proc Natl Acad Sci 94:13559–13564. https://doi.org/10.1073/pnas.94.25.13559
Zhukovsky E, Oprian D (1989) Effect of carboxylic acid side chains on the absorption maximum of visual pigments. Science 246:928–930. https://doi.org/10.1126/science.2573154
Acknowledgements
This work was by Ontario Graduate Scholarships (B.M.S., S.K.C.), a Vision Science Research Program Fellowship (S.K.C.), and an NSERC Discovery Grant (B.S.W.C.).
Author information
Authors and Affiliations
Contributions
Conceptualization: BMS, SKC, SGP, and BSWC; Methodology and investigation: BMS, SKC, and JL; Bioinformatics analyses: BMS and RKS; Visualization: AVN; Clinical variant data acquisition and curation: EH; Writing: SKC and BSWC, with input from BMS; Supervision: BSWC and SGP; All authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing Interests
All authors declare no competing interests.
Additional information
Handling editor: David Liberles.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Scott, B.M., Chen, S.K., Van Nynatten, A. et al. Scaling up Functional Analyses of the G Protein-Coupled Receptor Rhodopsin. J Mol Evol 92, 61–71 (2024). https://doi.org/10.1007/s00239-024-10154-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-024-10154-3