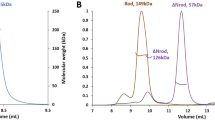
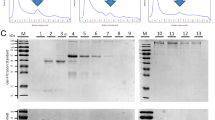
Abstract
Our previous calculations of ionic interactions indicated that the Caenorhabditis elegans intermediate filament (IF) IFA proteins, in addition to IFA/IFB-1 heterodimers, may also form homodimers. In order to prove the significance of these calculations, we analysed the dimerization potential of the IFA chains in blot overlays. Unexpectedly, we found here that the dimerization of the IFA-1 protein was of both homotypic and heterotypic nature, and involved all proteins immobilized on the membrane (IFA-1, IFA-2, IFA-4, IFB-1, IFB-2, IFC-1, IFC-2, IFD-1, IFD-2 and IFP-1). A similar interaction profile, though less complex, was observed for two biotinylated proteins (IFA-2 and IFA-4). These and previous results indicate that the IFA proteins are able to form many different heteropolymeric and homopolymeric complexes in the C. elegans tissue, but that only those triggered by the IFA-specific IFB-1 protein result in mature IFs. Moreover, the calculations of the possible ionic interactions between the individual rod sequences as well as their various deletion variants indicated a special role in this process for the middle part of the C. elegans IF coil 1B segment that is deleted in all vertebrate cytoplasmic IFs. We hypothesized here, therefore, that the striking promiscuity of the C. elegans IFs originally involved a nuclear lamin which, due to a two-heptad-long rod deletion, prevented formation of a functional lamin/cIF dimer. This, in concert with an efficient dimerization and a strict tissue-specific co-expression, may allow expansion and maintenance of the multiple Caenorhabditis IFs. A possible implication for evolution of chordate IFs proteins is also discussed.


Similar content being viewed by others
Abbreviations
- GFP:
-
Green fluorescent protein
- IF:
-
Intermediate filament
- RNAi:
-
RNA interference
References
Al-Hashimi H, Hall DH, Ackley BD, Lundquist EA, Buechner M (2018) Tubular excretory canal structure depends in intermediate filaments EXC-2 and IFA-4 in Caenorhabditis elegans. Genetics 210:637–652
Ben-Harush K, Wiesel N, Frenkiel-Krispin D, Moeller D, Soreq E, Aebi U, Herrmann H, Gruenbaum Y, Medalia O (2009) The supramolecular organization of the C. elegans nuclear lamin filament. J Mol Biol 386:392–1402
Blaxter M (2011) Nematodes: the worm and its relatives. PLoS Biol 9(4):1–9
Burkhard P, Kammerer RA, Steinmetz MO, Bourenkov GP, Aebi U (2000) The coiled-coil trigger site of the rod domain of cortexillin I unveils a distinct network of interhelical and intrahelical salt bridges. Structure 8:223–230
Carberry K, Wiesenfahrt T, Windoffer R, Bossinger O, Leube RE (2009) Intermediate filaments in Caenorhabditis elegans. Cell Motil Cytoskelet 66:852–864
Citi S, D’Atri F, Parry DAD (2000) Human and Xenopus cingulin a modular organization of the coiled-coil rod domain: predictions for intra- and intermolecular assembly. J Struct Biol 131:135–145
Coch RA, Leube RE (2016) Intermediate filaments and polarization in the intestinal epithelium. Cells 5:32. https://doi.org/10.3390/cells5030032
Dodemont H, Riemer D, Ledger N, Weber K (1994) Eight genes and alternative RNA processing pathways generate an unexpectedly large diversity of cytoplasmic intermediate filament proteins in the nematode Caenorhabditis elegans. EMBO J 13:2625–2638
Ehrlich F, Fischer H, Langbein L, Praetzel-Wunder S, Ebner B, Figlak K, Weissenbacher A, Sipos W, Tschachler E, Eckhart L (2019) Differential evolution of the epidermal keratin cytoskeleton in terrestrial and aquatic mammals. Mol Biol Evol 26(2):328–340
Erber A, Riemer D, Bovenschulte M, Weber K (1998) Molecular phylogeny of metazoan intermediate filament proteins. J Mol Evol 47:751–762
Erber A, Riemer D, Hofemeister H, Bovenschulte M, Stick R, Panopoulou G, Lehrach H, Weber K (1999) Characterisation of the Hydra lamin and its gene; a molecular phylogeny of metazoan lamins. J Mol Evol 49:260–271
Francis R, Waterston RH (1991) Muscle cell attachement in Caenorhabditis elegans. J Cell Biol 114:465–479
Fraser AG, Kamath RS, Zipperlen P, Martinez-Campos M, Sohrmann M, Ahringer J (2000) Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature 408:325–330
Fuchs E, Weber K (1994) Intermediate filaments: structure, dynamics, function and disease. Annu Rev Biochem 63:345–382
Geisler N, Schünemann J, Weber K, Häner M, Aebi U (1998) Assembly and architecture of invertebrate cytoplasmic intermediate filaments reconcile features of vertebrate cytoplasmic and nuclear lamin-type intermediate filaments. J Mol Biol 282:601–617
Geisler F, Gerhardus H, Carberry K, Davis W, Jorgensen E, Richardson C, Bossinger O, Leube RE (2016) A novel function for the MAP kinase SMA-5 in intestinal tube stability. Mol Biol Cell 27(24):3855–3868
Geisler F, Coch RA, Richardson C, Goldberg M, Denecke M, Bossinger O, Leube RE (2019) The intestinal intermediate filament network responds to and protects against microbial insults and toxins. Development 146(2):dev169482. https://doi.org/10.1242/dev.169482
Hapiak V, Hresko MC, Schriefer LA, Saiyasisongkhram K, Bercher M, Plenefisch JD (2003) mua-6, a gene required for tissue integrity in Caenorhabditis elegans, encodes a cytoplasmic intermediate filament. Dev Biol 263:330–342
Hatzfeld M, Franke WW (1985) Pair formation and promiscuity of cytokeratins: formation in vitro of heterotypic complexes and intermediate-sized filaments by homologous and heterologous recombinations of purified polypeptides. J Cell Biol 101:1826–1841
Herrmann H, Häner M, Brettel M, Müller S, Goldie K, Fedtke B, Lustig A, Franke WW, Aebi U (1996) Structure and assembly properties of the intermediate filament protein vimentin: the role of its head, rod and tail domains. J Mol Biol 264:933–953
Herrmann H, Strelkov SV, Burkhard P, Aebi U (2009) Intermediate filament: primary determinants of cell architecture and plasticity. J Clin Investig 119:1772–1783
Hesse M, Franz T, Tamai Y, Taketo MM, Magin TM (2000) Targeted deletion of keratin 18 and 19 leads to trophoblast fragility and early embryonic lethality. EMBO J 19:5060–5070
Hresko MC, Williams BD, Waterston RH (1994) Assembly of body wall muscle and muscle cell attachment structures in Caenorhabditis elegans. J Cell Biol 124:491–506
Hresko MC, Schriefer LA, Shrimankar P, Waterston RH (1999) Myotactin, a novel hypodermal protein involved in muscle-cell adhesion in Caenorhabditis elegans. J Cell Biol 146:659–672
Hüsken K, Wiesenfahrt T, Abraham Ch, Windoffer R, Bossinger O, Leube RE (2008) Maintenance of the intestinal tube in Caenorhabditis elegans: the role of the intermediate filament protein IFC-2. Differentiation 76:881–896
Irvine AD, McLean WH (1999) Human keratin diseases: the increasing spectrum of disease and subtlety of the phenotype-genotype correlation. Br J Dermatol 140:815–828
Karabinos A (2013) The cephalochordate Branchiostoma genome contains 26 intermediate filament (IF) genes: implication for evolution of chordate IF proteins. Eur J Cell Biol 92(8–9):295–302
Karabinos A (2019) Intermediate filament (IF) proteins IFA-1 and IFB-1 represent a basic heteropolymeric IF cytoskeleton of nematodes: a molecular phylogeny of nematode IFs. Gene 692:44–53
Karabinos A, Schmidt H, Harborth J, Schnabel R, Weber K (2001) Essential roles for four cytoplasmic intermediate filament proteins in Caenorhabditis elegans development. Proc Natl Acad Sci USA 98(14):7863–7868
Karabinos A, Schulze E, Klisch T, Wang J, Weber K (2002a) Expression profiles of the essential intermediate filament (IF) protein A2 and the IF protein C2 in the nematode Caenorhabditis elegans. Mech Dev 117:311–314
Karabinos A, Schünemann J, Parry DAD, Weber K (2002b) Tissue-specific coexpression and in vitro heteropolymer formation of the two small Branchiostoma intermediate filament proteins A3 and B2. J Mol Biol 316:127–137
Karabinos A, Schulze E, Schünemann J, Parry DAD, Weber K (2003a) In vivo and in vitro evidence that the four essential intermediate filament (IF) proteins A1, A2, A3 and B1 of the nematode Caenorhabditis elegans form an obligate heteropolymeric IF system. J Mol Biol 333:307–319
Karabinos A, Schünemann J, Meyer M, Aebi U, Weber K (2003b) The single nuclear lamin of Caenorhabditis elegans forms in vitro stable intermediate filaments and paracrystals with a reduced axial periodicity. J Mol Biol 325:241–247
Karabinos A, Schünemann J, Weber K (2004a) Most genes encoding cytoplasmic intermediate filament (IF) proteins of the nematode Caenorhabditis elegans are required in late embryogenesis. Eur J Cell Biol 83(9):457–468
Karabinos A, Zimek A, Weber K (2004b) The genome of the early chordate Ciona intestinalis encodes only five cytoplasmic intermediate filament proteins including a single type I and type II keratin and a unique IF-annexin fusion protein. Gene 326:123–129
Karabinos A, Schünemann J, Parry DAD (2012) A rod domain sequence in segment 1B triggers dimerisation of the two small Branchiostoma IF proteins B2 and A3. Eur J Cell Biol 91:800–808
Karabinos A, Schünemann J, Parry DAD (2017) Assembly studies of six intestinal intermediate filament (IF) proteins B2, C1, C2, D1, D2, and E1 in the nematode C. elegans. Cytoskeleton 74(3):107–113
Karabinos A, Schulze E, Baumeister R (2019) Analysis of the novel excretory cell expressed ECP-1 proteins and its proposed ECP-1/IFC-2 fusion proteins EXC-2 in the nematode Caenorhabditis elegans. Gene Expr Patterns 34:119061
Kolb T, Maaβ K, Hergt M, Aebi U, Herrmann H (2011) Lamin A and lamin C form homodimers and coexist in higher complex forms both in the nucleoplasmic fraction and in the lamina of cultured cells. Nucleus 2(5):425–433
Kollmar M (2015) Polyphyly of nuclear lamin genes indicates an early eukaryotic origin of the metazoan-type intermediate filament proteins. Sci Rep 5:10652. https://doi.org/10.1038/srep10652
Kolotuev I, Hyenne V, Schwab Y, Rodriguez Y, Labouesse M (2013) A pathway for unicellular tube extension depending on the lymphatic vessel determinant Prox1 and on osmoregulation. Nat Cell Biol 15(2):157–168
Liu J, Ben-Shahar TR, Riemer D, Treinin M, Spann P, Weber K, Fire A, Gruenbaum Y (2000) Essential roles for Caenorhabditis elegans lamin gene in nuclear organization, cell cycle progression, and spatial organization of nuclear pore complexes. Mol Biol Cell 11:3937–3947
Melcer S, Gruenbaum Y, Krohne G (2007) Invertebrate lamins. Exp Cell Res 313:2157–2166
Osmanagic-Myers S, Dechat T, Foisner R (2016) Lamins at the crossroads of mechanosignaling. Genes Dev 29:225–237
Parry DAD, Steinert PM (1995) Intermediate filament structure. Springer, New York
Peter A, Stick R (2015) Evolutionary aspects in intermediate filament proteins. Curr Opin Cell Biol 32:48–55
Suzuki KT, Suzuki M, Shigeta M, Fortriede JD, Takahashi S, Mawaribuchi S, Yamamoto T, Taira M, Fukui A (2017) Clustered Xenopus keratin genes: a genomic, transcriptomic, and proteomic analysis. Dev Biol 426:384–392
Vandebergh W, Bossuyt F (2012) Radiation and functional diversification of alpha keratins during early vertebrate evolution. Mol Biol Evol 29:995–1004
Vijayaraj P, Kröger K, Reuter U, Windoffer R, Leube RE, Magin TM (2009) Keratins regulate protein biosynthesis through localization of GLUT1 and -3 upstream of AMP kinase and Raptor. J Cell Biol 187:175–184
Wang J, Karabinos A, Schünemann J, Riemer D, Weber K (2000) The epidermal intermediate filament proteins of tunicates are distant keratins; a polymerisation-competent hetero coiled coil of the Styela D protein and Xenopus keratin 8. Eur J Cell Biol 79:478–487
Woo W-M, Goncharov A, Jin Y, Chisholm AD (2004) Intermediate filaments are required for C. elegans epidermal elongation. Dev Biol 267:216–229
Zhang H, Landmann F, Zahreddine H, Rodriguez D, Koch M, Labouesse M (2011) A tension-induced mechanotransduction pathway promotes epithelial morphogenesis. Nature 471:99–103
Acknowledgements
This work was funded by the European Regional Development OPVaV-2009/2.2/05-SORO (ITMS code:26220220143).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Authors proclaim no conflict of interests.
Additional information
Handling Editor: Kerry Geiler-Samerotte.
Electronic supplementary material
Below is the link to the electronic supplementary material.
239_2019_9904_MOESM1_ESM.doc
Fig. S1. (A) Alignment of the rod amino acid sequences of the C. elegans lamin (LMN-1) and cytoplasmic proteins IFA-1 to IFA-4, IFB-1, IFB-2, IFC-1, IFC-2, IFD-1, IFD-2 and IFP-1. Arrows pointing downwards and upwards mark the beginning and the end of the rod segments 1A, 1B and 2. The linkers L1 and L12, as well as the stutter in coil 2 are also indicated. Asterisks between the rod segments of the individual proteins mark the a and d positions of the heptad repeat pattern. Dashes are used to optimize the sequence alignment. Bold letters indicate residues that are identical in all twelve proteins. The 42 residue-long region of the C. elegans IF coil 1B segments, which is deleted by all chordate short/S-type IF proteins, is marked by dots above the sequences. The vertical lines between the individual sequences mark the positions of the charged residues in the corresponding coil segments which are involved in potential 2e′-1 g, 1 g′-2e, 2a′-1 g, 1 g′-2a, 2e′-1d, and 1d′-2e attractive (normal lines) or repulsive (dashed lines) salt bridges (see text for details). (B) Alignment of the full-length (fl) C. elegans Lamin (CeLamin_fl) and the seventeen nematode Lamins (the presented alignment was retrieved from the CyMoBase; Kollmar 2015). The arrowheads pointing downwards and upwards mark the beginning and the end of the rod segments 1A, 1B and 2, which are connected by linkers L1 and L12, respectively (for details, see the text). Dashes are used to optimize the sequence alignment. Note that only C. elegans Lamin harbours 14 deleted residues in the coil 2B but the same deletion is also seen in the related Caenorhabditis remanei, Caenorhabditis brenneri and Caenorhabditis briggsae species (data not shown). Also note that Lamins from another two clade V Heterorhabditis and Oscheius species have a coil 2 which is only one heptad shorter. The compared full-length (fl) Lamin sequences are derived from the following nematode species: Clade I: Trichinella spiralis (Trs), Trichuris muris (Trm); Clade III: Ascaris suum (Ass), Brugia malayi (Brm), Onchocerca volvulus (Ov), Wuchereria bancrofti (Wb); Clade IV: Bursaphelenchus xylophilus (Bx), Globodera pallida (Glp), Heterodera glycines (Heg), Meloidogyne incognita (Mi), Meloidogyne hapla (Mh), Panagrellus redivivus (Par); Clade V: Caenorhabditis elegans (Ce), Heterorhabditis baderophora (Hb), Oscheius tipulae (Oct), Pristionchus pacificus (Psp). The clades III (Spirurina), IV (Tylenchina and other) and V (Rhabditina) belong to Chromadoria, which diverged during nematode evolution from the more ancient Dorylaimia (clade I) and Enoplia (clade II; for review see Blaxter, 2011). Supplementary material 1 (DOC 288 kb)
Rights and permissions
About this article
Cite this article
Karabinos, A., Schünemann, J. & Parry, D.A.D. Promiscuous Dimerization Between the Caenorhabditis elegans IF Proteins and a Hypothesis to Explain How Multiple IFs Persist Over Evolutionary Time. J Mol Evol 87, 221–230 (2019). https://doi.org/10.1007/s00239-019-09904-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-019-09904-5