Abstract
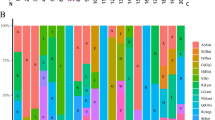
The Aux/IAA (IAA) gene family, involved in the auxin signalling pathway, acts as an important regulator in plant growth and development. In this study, we explored the evolutionary trajectory of the IAA family in common wheat. The results showed ten pairs of paralogs among 34 TaIAA family members. Seven of the pairs might have undergone segmental duplication, and the other three pairs appear to have experienced tandem duplication. Except for TaIAA15-16, these duplication events occurred in the ancestral genomes before the divergence of Triticeae. After that point, two polyploidization events shaped the current TaIAA family consisting of three subgenomic copies. The structure or expression pattern of the TaIAA family begins to differentiate in the hexaploid genome, where TaIAAs in the D genome lost more genes (eight) and protein secondary structures (α1, α3 and β5) than did the other two genomes. Expression analysis showed that six members of the TaIAA family were not expressed, and members such as TaIAA8, 15, 16, 28 and 33 exhibited tissue-specific expression patterns. In addition, three of the ten pairs of paralogs (TaIAA5–12, TaIAA15–16 and TaIAA29–30) showed similar expression patterns, and another five paralog pairs displayed differential expression patterns. Phylogenetic analysis showed that paralog pairs with high rates of evolution (ω > ω 0), particularly TaIAA15–16 and TaIAA29–30, experienced greater motif loss, with only zero to two interacting IAA proteins. In contrast, most paralogous genes with low ω, such as TaIAA5–12, had more complete motifs and higher degrees of interaction with other family members.






Similar content being viewed by others
References
Abel S, Theologis A (1995) A polymorphic bipartite motif signals nuclear targeting of early auxin-inducible proteins related to PS-IAA4 from pea (Pisum sativum). Plant J 8:87–96
Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS (2009) MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37:W202-208
Birchler JA, Veitia RA (2007) The gene balance hypothesis: from classical genetics to modern genomics. Plant Cell 19:395–402
Brassica rapa Genome Sequencing Project Consortium (2011) The genome of the mesopolyploid crop species Brassica rapa. Nat Genet 43:1035–1039
Brenchley R, Spannagl M, Pfeifer M, Barker GL, D’Amore R, Allen AM, McKenzie N, Kramer M, Kerhornou A, Bolser D, Kay S, Waite D, Trick M, Bancroft I, Gu Y, Huo N, Luo MC, Sehgal S, Gill B, Kianian S, Anderson O, Kersey P, Dvorak J, McCombie WR, Hall A, Mayer KF, Edwards KJ, Bevan MW, Hall N (2012) Analysis of the bread wheat genome using whole-genome shotgun sequencing. Nature 491:705–710
Buggs RJ, Zhang L, Miles N, Tate JA, Gao L, Wei W, Schnable PS, Barbazuk WB, Soltis PS, Soltis DE (2011) Transcriptomic shock generates evolutionary novelty in a newly formed, natural allopolyploid plant. Curr Biol 21:551–556
Chen ZJ (2007) Genetic and epigenetic mechanisms for gene expression and phenotypic variation in plant polyploids. Annu Rev Plant Biol 58:377–406
Comai L (2005) The advantages and disadvantages of being polyploid. Nat Rev Genet 6:836–846
Cooke TJ, Poli D, Sztein AE, Cohen JD (2002) Evolutionary patterns in auxin action. Plant Mol Biol 49:319–338
D’Hont A, Denoeud F, Aury JM (2012) The banana (Musa acuminata) genome and the evolution of monocotyledonous plants. Nature 488:213–217
De Smet I, Voss U, Lau S, Wilson M, Shao N, Timme RE, Swarup R, Kerr I, Hodgman C, Bock R, Bennett M, Jürgens G, Beeckman T (2011) Unraveling the evolution of auxin signaling. Plant Physiol 155:209–221
Dinesh DC, Kovermann M, Gopalswamy M, Hellmuth A, Calderón Villalobos LI, Lilie H, Balbach J, Abel S (2015) Solution structure of the PsIAA4 oligomerization domain reveals interaction modes for transcription factors in early auxin response. Proc Natl Acad Sci USA 112:6230–6235
Errami M, Geourjon C, Deléage G (2003) Detection of unrelated proteins in sequences multiple alignments by using predicted secondary structures. Bioinformatics 19:506–512
Freeling M, Thomas BC (2006) Gene-balanced duplications, like tetraploidy, provide predictable drive to increase morphological complexity. Genome Res 16:805–814
Gaut BS, Morton BR, McCaig BC, Clegg MT (1996) Substitution rate comparisons between grasses and palms: synonymous rate differences at the nuclear gene Adh parallel rate differences at the plastid gene rbcL. Proc Natl Acad Sci USA 93:10274–10279
Guo Y, Qiu LJ (2013) Genome-wide analysis of the Dof transcription factor gene family reveals soybean-specific duplicable and functional characteristics. PLoS ONE 8:e76809
Halliday KJ, Martínez-García JF, Josse EM (2009) Integration of light and auxin signaling. Cold Spring Harb Perspect Biol 1:a001586
Huang Z, Duan W, Song X, Tang J, Wu P, Zhang B, Hou X (2015) Retention, molecular evolution, and expression divergence of the auxin/indole acetic acid and auxin response factor gene families in Brassica rapa shed light on their evolution patterns in plants. Genome Biol Evol 8:302–316
Ihaka R, Gentleman R (1996) R: a language for data analysis and graphics. J Comput Graph Stat 5:299–314
International Wheat Genome Sequencing Consortium (IWGSC) (2014) A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 345:1251788
Jing H, Yang X, Zhang J, Liu X, Zheng H, Dong G, Nian J, Feng J, Xia B, Qian Q, Li J, Zuo J (2015) Peptidyl-prolyl isomerization targets rice Aux/IAAs for proteasomal degradation during auxin signalling. Nat Commun 6:7395
Jung H, Lee DK, Choi YD, Kim JK (2015) OsIAA6, a member of the rice Aux/IAA gene family, is involved in drought tolerance and tiller outgrowth. Plant Sci 236:304–312
Kazan K, Manners JM (2009) Linking development to defense: auxin in plant-pathogen interactions. Trends Plant Sci 14:373–382
Koonin EV (2005) Orthologs, paralogs, and evolutionary genomics. Annu Rev Genet 39:309–338
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948
Leitch A, Leitch I (2008) Genomic plasticity and the diversity of polyploid plants. Science 320:481–483
Letunic I, Doerks T, Bork P (2015) SMART: recent updates, new developments and status in 2015. Nucleic Acids Res 43:257–260
Li WH (1997) Molecular evolution. Sinauer Associates, Sunderland
Liu B, Vega JM, Feldman M (1998) Rapid genomic changes in newly synthesized amphiploids of Triticum and Aegilops II Changes in low-copy coding DNA sequences. Genome 41:535–542
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆CT method. Methods 25:402–408
Lynch M, Conery JS (2000) The evolutionary fate and consequences of duplicate gene. Science 290:1151–1155
Masterson J (1994) Stomatal size in fossil plants: evidence for polyploidy in majority of angiosperms. Science 264:421–424
Michael TP, VanBuren R (2015) Progress, challenges and the future of crop genomes. Curr Opin Plant Biol 24:71–81
Nanao MH, Vinos-Poyo T, Brunoud G, Thévenon E, Mazzoleni M, Mast D, Lainé S, Wang S, Hagen G, Li H, Guilfoyle TJ, Parcy F, Vernoux T, Dumas R (2014) Structural basis for oligomerization of auxin transcriptional regulators. Nat Commun 5:3617
Nystedt B, Street NR, Wetterbom A (2013) The Norway spruce genome sequence and conifer genome evolution. Nature 497:579–584
Otto SP (2007) The evolutionary consequences of polyploidy. Cell 131:452–462
Paponov IA, Teale W, Lang D, Paponov M, Reski R, Rensing SA, Palme K (2009) The evolution of nuclear auxin signalling. BMC Evol Biol 9:126
Petersen G, Seberg O, Yde M, Berthelsen K (2006) Phylogenetic relationships of Triticum and Aegilops and evidence for the origin of the A, B, and D genomes of common wheat (Triticum aestivum). Mol Phylogenet Evol 39:70–82
Pfeifer M, Kugler KG, Sandve SR, Zhan B, Rudi H, Hvidsten TR, International Wheat Genome Sequencing Consortium, Mayer KF, Olsen OA (2014) Genome interplay in the grain transcriptome of hexaploid bread wheat. Science 345:1250091
Pont C, Murat F, Guizard S, Flores R, Foucrier S, Bidet Y, Quraishi UM, Alaux M, Doležel J, Fahima T, Budak H, Keller B, Salvi S, Maccaferri M, Steinbach D, Feuillet C, Quesneville H, Salse J (2013) Wheat syntenome unveils new evidences of contrasted evolutionary plasticity between paleo- and neoduplicated subgenomes. Plant J 76:1030–1044
Priya R, Ive DS (2013) Evolutionary aspects of auxin signalling. In: Zažímalová E (ed) Auxin and its role in plant development, 1st edn. Springer, Vienna, pp 265–290
Qiao L, Zhang X, Han X, Zhang L, Li X, Zhan H, Ma J, Luo P, Zhang W, Cui L, Li X, Chang Z (2015) A genome-wide analysis of the auxin/indole-3-acetic acid gene family in hexaploid bread wheat (Triticum aestivum L). Front Plant Sci 6:770
Raes J, Van de Peer Y (2003) Gene duplications, the evolution of novel gene functions, and detecting functional divergence of duplicates in silico. Appl Bioinformatics 2:92–101
Roulin A, Auer PL, Libault M, Schlueter J, Farmer A, May G, Stacey G, Doerge RW, Jackson SA (2013) The fate of duplicated genes in a polyploid plant genome. Plant J 73:143–153
Salse J, Bolot S, Throude M, Jouffe V, Piegu B, Quraishi UM, Calcagno T, Cooke R, Delseny M, Feuillet C (2008) Identification and characterization of shared duplications between rice and wheat provide new insight into grass genome evolution. Plant Cell 20:11–24
Schaller GE, Bishopp A, Kieber JJ (2015) The Yin-Yang of hormones: cytokinin and auxin interactions in plant development. Plant Cell 27:44–63
Singla B, Chugh A, Khurana JP, Khurana P (2006) An early auxin-responsive Aux/IAA gene from wheat (Triticum aestivum) is induced by epibrassinolide and differentially regulated by light and calcium. J Exp Bot 57:4059–4070
Soltis PS, Douglas E. Soltis DE, Savolainen V, Crane PR, Barraclough TG (2002) Rate heterogeneity among lineages of tracheophytes: integration of molecular and fossil data and evidence for molecular living fossils. Proc Natl Acad Sci USA 99:4430–4435
Song Y, You J, Xiong L (2009) Characterization of OsIAA1 gene, a member of rice Aux/IAA family involved in auxin and brassinosteroid hormone responses and plant morphogenesis. Plant Mol Biol 70:297–309
Strader LC, Chen GL, Bartel B (2010) Ethylene directs auxin to control root cell expansion. Plant J 64:874–884
Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, Kuhn M, Bork P, Jensen LJ, von Mering C (2015) STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 43:D447-452
Talboys PJ, Healey JR, Withers PJ, Jones DL (2014) Phosphate depletion modulates auxin transport in Triticum aestivum leading to altered root branching. J Exp Bot 65:5023–5032
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Thakur JK, Jain M, Tyagi AK, Khurana JP (2005) Exogenous auxin enhances the degradation of a light down-regulated and nuclear-localized OsiIAA1, an Aux/IAA protein from rice, via proteasome. Biochim Biophys Acta 1730:196–205
Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ (2009) Jalview Version 2: a multiple sequence alignment editor and analysis workbench. Bioinformatics 25:1189–1191
Wheeler TJ, Eddy SR (2013) nhmmer: DNA homology search with profile HMMs. Bioinformatics 29:2487–2489
Winkler M, Niemeyer M, Hellmuth A, Janitza P, Christ G, Samodelov SL, Wilde V, Majovsky P, Trujillo M, Zurbriggen MD, Hoehenwarter W, Quint M, Calderón Villalobos LIA (2017) Variation in auxin sensing guides AUX/IAA transcriptional repressor ubiquitylation and destruction. Nat Commun 8:15706
Yang Z (1997) PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci 13:555–556
Yasumura Y, Crumptontaylor M, Fuentes S, Harberd NP (2007) Step-by-step acquisition of the gibberellin-DELLA growth-regulatory mechanism during land-plant evolution. Curr Biol 17:1225–1230
Zhang H, Zhu B, Qi B, Gou X, Dong Y, Xu C, Zhang B, Huang W, Liu C, Wang X, Yang C, Zhou H, Kashkush K, Feldman M, Wendel JF, Liu B (2014) Evolution of the BBAA component of bread wheat during its history at the allohexaploid level. Plant Cell 26:2761–2776
Acknowledgements
This study was funded by the National Key Research and Development Plan of China (2016YFD0102004-07), the National Natural Science Foundation of China (31601307), Shanxi Province Science Foundation for Youths (2015021145), Shanxi Province Natural Science Foundation (201601D102051) and Shanxi Province International Cooperation Project (201603D421003). We thank Dr. Xiaoyan Li (Beijing Anzhen Hospital Affiliated to the Capital Medical University) and Dr. Wenping Zhang (Fujian Agriculture and Forestry University) for their assistance in the RT-PCR experiment.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Qiao, L., Zhang, L., Zhang, X. et al. Evolution of the Aux/IAA Gene Family in Hexaploid Wheat. J Mol Evol 85, 107–119 (2017). https://doi.org/10.1007/s00239-017-9810-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-017-9810-z