Abstract
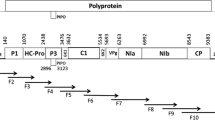
RNA-dependent RNA polymerase (RdRp) encoded by ORF2 and putative aphid transmission factor (PATF) encoded by ORF5 of Sugarcane yellow leaf virus (SCYLV) were detected in six sugarcane cultivars affected by yellow leaf using RT-PCR and real-time RT-PCR assays. Expression of both genes varied among infected plants, but overall expression of RdRp was higher than expression of PATF. Cultivar H87-4094 from Hawaii yielded the highest transcript levels of RdRp, whereas cultivar C1051-73 from Cuba exhibited the lowest levels. Sequence comparisons among 25 SCYLV isolates from various geographical locations revealed an amino acid similarity of 72.1–99.4 and 84.7–99.8 % for the RdRp and PATF genes, respectively. The 25 SCYLV isolates were separated into three (RdRp) and two (PATF) phylogenetic groups using the MEGA6 program that does not account for genetic recombination. However, the SCYLV genome contained potential recombination signals in the RdRp and PATF coding genes based on the GARD genetic algorithm. Use of this later program resulted in the reconstruction of phylogenies on the left as well as on the right sides of the putative recombination breaking points, and the 25 SCYLV isolates were distributed into three distinct phylogenetic groups based on either RdRp or PATF sequences. As a result, recombination reshuffled the affiliation of the accessions to the different clusters. Analysis of selection pressures exerted on RdRp and PATF encoded proteins revealed that ORF 2 and ORF 5 underwent predominantly purifying selection. However, a few sites were also under positive selection as assessed by various models such as FEL, IFEL, REL, FUBAR, MEME, GA-Branch, and PRIME.







Similar content being viewed by others
References
Aaziz R, Tepfer M (1999) Recombination in RNA viruses and in virus-resistant transgenic plants. J Gen Virol 80:1339–1346
Abu Ahmad Y, Royer M, Daugrois JH, Costet L, Lett JM, Victoria JI, Girard JC, Rott P (2006) Geographical distribution of four Sugarcane yellow leaf virus genotypes. Plant Dis 90:1156–1160
Abu Ahmad Y, Costet L, Daugrois J-H, Nibouche S, Letourmy P, Girard J-C, Rott P (2007) Variation in infection capacity and in virulence exists between genotypes of Sugarcane yellow leaf virus. Plant Dis 91:253–259
Akaike H (1974) A new look at the statistical model identification. IEEE Trans Autom Control 19:716–723
Aljanabi S, Parmessur Y, Moutia Y, Saumtally S, Dookun A (2001) Further evidence of the association of a phytoplasma and a virus with yellow leaf syndrome in sugarcane. Plant Path 50:628–636
Atchley WR, Zhao J, Fernandes AD, Druke T (2005) Solving the protein sequence metric problem. PNAS 102:6395–6400
Boulila M (2011) Selective constraints, molecular recombination structure and phylogenetic reconstruction of isometric plant RNA viruses of the families Luteoviridae and Tymoviridae. Biochimie 93:242–253
Chao L (1988) Evolution of sex in RNA viruses. J Theor Biol 133:99–112
Chao L (1997) Evolution of sex and the molecular clock in RNA viruses. Gene 205:301–308
Chare ER, Holmes EC (2006) A phylogenetic survey of recombination frequency in plant RNA viruses. Arch Virol 15:933–946
Chay C, Smith DM, Vaughan R, Gray SM (1996) Diversity among isolates within the PAV serotype of barley yellow dwarf virus. Phytopathology 86:370–377
Chinnaraja C, Viswanathan R, Karuppaiah R, Bagyalakshmi K, Malathi P, Parameswari B (2013) Complete genome characterization of Sugarcane yellow leaf virus from India: Evidence for RNA recombination. Eur J Plant Pathol 135:335–349
Conant GC, Wagner GP, Stadler PF (2007) Modeling amino acid substitution pattern in orthologous genes. Mol Phyl Evol 42:298–307
Corpet F (1988) Multiple sequence alignment with hierarchical clustering. Nucl Acids Res 16:10881–10890
Cronjǐe P, Tymon A, Jones P, Bailey R (1998) Association of a phytoplasma with a yellow leaf syndrome of sugarcane in Africa. Ann Appl Biol 133:177–186
Delport W, Poon AFY, Frost SDW, Kosakovsky Pond SL (2010) Datamonkey 2010: a suite of phylogenetic analysis tools for evolutionary biology. Bioinformatics 29:2455–2457
Domingo E, Holland JJ (1997) RNA virus mutations and fitness for survival. Ann Rev Microb 51:151–178
ElSayed AI (2013) Maize (Zea mays L.) constitutes a novel host to Sugarcane yellow leaf virus. Cana J Plant Pathol 35:68–74
ElSayed AI, Komor E (2012) Investigation of ORF0 as a sensitive alternative diagnostic segment to detect Sugarcane yellow leaf virus. J Gen Plant Path 78:207–216
ElSayed AI, Komor E (2013) Quantitative analysis of transcripts of the open reading frames of Sugarcane yellow leaf virus genome by one-multiplex RT-PCR: evidence for a high transcript level of suppressor gene in sink leaves. J Phytopathol 161:774–783
ElSayed AI, Weig A, Komor E (2011) Molecular characterization of Hawaiian Sugarcane yellow leaf virus genotypes and their phylogenetic relationship to strains from other sugarcane-growing countries. Eur J Plant Pathol 129:399–412
ElSayed AI, Boulila M, Komor E, Zhu YJ (2012) Putative recombination signature and significance of deletion/insertion events in RdRp coding region of Sugarcane yellow leaf virus. Biochimie 94:1764–1772
Haenni A-L (2008) In: Roossinck MJ (ed) Virus evolution and taxonomy, plant virus evolution. Springer, Berlin, pp 205–217
Khishino H, Hasegawa M (1989) Evaluation of the maximum likelihood estimate of the evolutionary tree topologies from DNA sequence data, and the branching order in Hominoidea. J Mol Evol 29:170–179
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Korber B (2000) HIV signatures and similarities. In: Rodrigo AG, Learn GH Jr (eds) Computational and evolutionary analysis of HIV molecular sequences. Kluwer Academic, Dordrecht, pp 55–72
Kosakovsky Pond SL, Frost SDW (2005a) Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics 21:2531–2533
Kosakovsky Pond SL, Frost SDW (2005b) A genetic algorithm approach to detecting lineage-specific variation in selection pressure. Mol Biol Evol 22:478–485
Kosakovsky Pond SL, Frost SDW, Muse SV (2005) HyPhy: hypothesis testing using phylogenies. Bioinformatics 21:676–679
Kosakovsky Pond SL, Posada D, Gravenor MB, Woelk CH, Frost SDW (2006a) GARD: a genetic algorithm for recombination detection. Bioinformatics 22:3096–3098
Kosakovsky Pond SL, Posada D, Gravenor MB, Woelk CH, Frost SDW (2006b) Automated phylogenetic detection of recombination using a genetic algorithm. Mol Biol Evol 23:1891–1901
Kosakovsky Pond SL, Frost SD, Grossman Z, Gravenor MB, Richman DD, Brown AJ (2006c) Adaptation to different human populations by HIV-1 revealed by codon-based analyses. PLoS Comput Biol 2:530–538
Larkin MA, Blackshileds G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948
Lehrer AT, Komor E (2008) Symptom expression of yellow leaf disease in sugarcane cultivars with different degrees of infection by Sugarcane yellow leaf virus. Plant Pathol 57:178–189
Lin YH, Gao SJ, Damaj MB, Fu HY, Chen RK, Mirkov TE (2014) Genome characterization of Sugarcane yellow leaf virus from China reveals a novel recombinant genotype. Arch Virol. doi:10.1007/s00705-013-1957-3
Lockhart BEL, Cronjǐe CPR (2000) Yellow leaf syndrome. In: Rott P, Bailey RA, Comstock JC, Croft BJ, Saumtally AS (eds) A guide to sugarcane diseases. CIRAD-ISSCT, Montpellier, France, pp 291–295
Martin RR, Keese PK, Young JM, Waterhouse PM, Gerlach WL (1990) Evolution and molecular biology of luteoviruses. Ann Rev Phytopathol 28:341–363
Moonan F, Molina J, Mirkov TE (2000) Sugarcane yellow leaf virus: an emerging virus that has evolved by recombination between luteoviral and poleroviral ancestors. Virology 269:156–171
Murrell B, Wertheim JO, Moola S, Weighill T, Scheffler K, Kosakovsky Pond SL (2012) Detecting individual sites subject to episodic diversifying selection. PLoS Genet 8:e1002764. doi:10.1371/journal.pgen.1002764
Murrell B, Moola S, Mabona A, Weighill T, Sheward D, Kosakovsky Pond SL, Scheffler K (2013) FUBAR: A Fast, Unconstrained Bayesian AppRoximation for inferring selection. Mol Biol Evol 30:1196–1205
Nagai M, Sakoda Y, Mori M, Hayashi M, Kida H, Akashi H (2003) Insertion of a cellular sequence and RNA recombination in the structural protein coding region of cytopathogenic bovine viral diarrhea virus. J Gen Virol 84:447–452
Ohta T (1973) Slightly deleterious mutant substitutions in evolution. Nature 246:96–98
Ohta T (1974) Mutational pressure as the main cause of molecular evolution and polymorphism. Nature 252(29):351–354
Ohta T (1992) The nearly neutral theory of molecular evolution. Ann Rev Ecol Evol Syst 23:263–286
Pathak KB, Nagy PD (2009) Defective interfering RNAs: foes of viruses and friends of virologists. Viruses 1:895–919
Power AG (2000) Insect transmission of plant viruses: a constraint on virus variability. Curr Opin Plant Biol 3:336–340
Reanney DC (1982) The evolution of RNA viruses. Ann Rev Microb 13:47–73
Roossinck JM (1997) Mechanisms of plant virus evolution. Ann Rev Phytopathol 35:191–209
Rott P, Mirkov TE, Schenck S, Girard J-C (2007) Recent advances in research on Sugarcane yellow leaf virus, the causal agent of sugarcane yellow leaf. In: Proceedings international society of sugarcane technologists congress 26 (CD Rom).
Scheffler K, Martin DP, Seoighe C (2006) Robust inference of positive selection from recombining coding sequences. Bioinformatics 22:2493–2499
Smith GR, Borg Z, Lockhart BEL, Braithwaite KS, Gibbs MJ (2000) Sugarcane yellow leaf virus: a novel member of the Luteoviridae that probably arose by inter-species recombination. J Gen Virol 81:1865–1869
Tajima F (1989) Statistical-method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585–595
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729
Viswanathan R (2004) Ratoon stunting disease infection favours severity of yellow leaf syndrome caused by Sugarcane yellow leaf virus in sugarcane. Sugarcane Inter 22:3–7
Viswanathan R, Balamuralikrishnan M, Karuppaiah R (2008) Identification of three genotypes of Sugarcane yellow leaf virus causing yellow leaf disease from India and their molecular characterization. Virus Gen 37:368–379
Wang MQ, Zhou GH (2010) A near complete genome sequence of a distinct isolate of Sugarcane yellow leaf virus from China, representing a sixth new genotype. Virus Gen 41:268–272
Wang MQ, Xu DL, Li R, Zhou GH (2012) Genotype identification and genetic diversity of Sugarcane yellow leaf virus in China. Plant Pathol 61:986–993
Worobey M, Holmes EC (1999) Evolutionary aspects of recombination in RNA viruses. J Gen Virol 80:2535–2543
Zaccomer B, Haenni AL, Macaya G (1995) The remarkable variety of plant RNA virus genomes. J Gen Virol 76:231–247
Acknowledgments
The authors are grateful to Dr. Axel Lehrer (Hawaiian Agriculture Research Center, Aiea, USA) and to Dr. Isabel Medina Borges (Havana, Cuba) for the gift of sugarcane cultivars. This work was supported by the research fund from Zagazig University, Egypt.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
ElSayed, A.I., Boulila, M. & Rott, P. Molecular Evolutionary History of Sugarcane yellow leaf virus Based on Sequence Analysis of RNA-Dependent RNA Polymerase and Putative Aphid Transmission Factor-Coding Genes. J Mol Evol 78, 349–365 (2014). https://doi.org/10.1007/s00239-014-9630-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-014-9630-3