Abstract
Calotropis procera produces a latex used in traditional medicine because of its components which are found to be biologically active. The latex of C. procera has many benefits such as pain reduction, antimicrobial attitude and other precious attributes. Latex lysozyme (LL) is a particular protein contained in C. procera plant having a promising antibacterial property. The process applied for extraction implicated ammonium sulfate precipitation, water extraction, ion exchange and gel filtration column chromatography evidencing 14.4 kDa for LL as a molecular mass. LL after purification showed 397.18 U mg−1 as specific activity (10.30 purification fold), and with 37.9% as a yield. In addition, the chemical and physical analyses showed that LL had its best realization when the pH was 5.5 at 50 °C. Moreover when metal ions such as Ca2+ and Mg2+ were incorporated the relative activity of the enzyme resulted ampliflied. When a purification was conducted with CM-Cellulose and Sephadex G-100 chromatography, LL revealed an antibacterial property versus both Escherichia coli (MICs 14 µg ml−1) and Bacillus cereus (MICs 13 µg ml−1). HR-TEM analysis displayed an antimicrobial potential of LL after its purification; that analysis showed the deterioration of the cell wall (external membrane of the cell) of both E. coli and B. cereus. Therefore, the LL obtained from C. procera maintains ability as an antibacterial enzyme applicable in various domains such as biotechnological and pharmaceutical industries.
Graphical abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
More than 20,000 species of plants distributed on over 40 angiosperm families (Papaveraceae, Apocynaceae, Asteraceae and Euphorbiaceae) contain latex. It is noteworthy that latex is produced by different families and orders of plants (not strictly related) displaying independent evolution [1,2,3,4]. Latex resides in the cytoplasm of laticifers which are typical cells with an elongated shape and distributed in leaves, stems and roots. Latex is composed by the laticifer vacuoles’ content which form an emulsion comprising: gum, tannins, oils, sugars, proteins, alkaloids, starches and terpenoids [2, 5, 6]. If plants are mechanically injured, for example for insect bites or for ruptures caused by wind, latex comes out and solidified when exposed to air [6].
The lacking in a shared operative mechanism is due to the different plants source of latex. Some proteins (pathogenesis-related and proteases) contained in the latex are constantly synthesized, but other proteins are produced in response to a stressful condition or pathogens’ attacks and wounds. Also the plant species can influence the latex proteomic and chemical composition [7,8,9]. C. procera (Apocynaceae family) widely grown in India. Its latex is composed by resinous compounds and cis or trans-polyisoprenes and contains a mixture of primary and specialized metabolites together with proteins and polypeptides. The latex produced by C. procera is used in India in traditional medicine to contrast the skin injuries, infestation by intestinal worms and food disorders [10, 11].
In African Countries, the latex obtained from the branches and roots of Calotropis procera is used in the traditional medicine against: leprosy, fever, ringworm, diarrhea, asthma, cough, seizures and eczema [12]. Pathogenesis-related proteins such as lysozyme are produced by various latex-producing plants to contrast pathogens injuries or attacks caused by pathogens [8, 13]. Lysozyme (crucial for self-defense) was accidentally discovered and is widely present in nature [14]. Lysozymes are known for their ability to extend antibacterial effects by employing muramidase to uniquely hydrolyze N-acetylmuramic acid 1,4 ß D linkage and peptidoglycan N-acetyl glucosamine [15, 16]. Antimicrobial proteins can be found in various plant tissues such as seeds, roots, leaves, flowers, fruits, and latices [17, 18]. A variety of antimicrobial proteins have been researched and characterized within the latex of plants like Hevea brasiliensis [19, 20], Carica papaya [21], and Asclepias syriaca [22, 23], all belonging to the lactiferous Apocynaceae family primarily found in tropical and subtropical regions. Although this plant exudate latex fluid from green leaves, shoot apex, and bark is commonly used in traditional medicine, there is limited available information regarding lysozyme enzymes sourced from C. procera latex.
In our current study, our objective was to extract and profile the lysozyme found within the latex of C. procera. We employed chromatographic methods to purify this latex lysozyme (LL), and subsequently assessed its antibacterial effectiveness against pathogenic bacteria.
Materials and methods
An assemblage of plant species and microbial cultures
Latex-producing plants were gathered from regions in and around Chennai, located in the Tamil Nadu region of India. The experimental microbial cultures were obtained from the Microbial Type Culture Collection & Gene Bank (MTCC) in Chandigarh, India.
Examination of lysozyme derived from latex-producing plants
The latex extracted from various plant species, including Achras sapota (Sapotaceae), Artrocarpus heterophyllus (Moraceae), Azadircachta indica (Meliaceae), Calotropis procera (Apocynaceae), Euphorbia tithymaloides (Euphorbiaceae), Ficus benghalensis (Moraceae), Ficus racemosa (Moraceae), Ficus religiosa (Moraceae), Ipomoea carnea (Convolvulaceae), Mangifera indica (Anacardiaceae), Musa acuminata (Musaceae), Nerium indicum (Apocynaaceae), Plumeria alba (Apocynaaceae), and Theivetia neriifolia (Apocynaaceae), was collected from both cultivated and uncultivated areas in Chennai, Tamil Nadu, India. The fresh latex was obtained from mature, healthy plants by making a superficial incision near the tender parts of leaves, and the latex was then drawn into a sterile glass beaker containing glass-distilled water in a 1:1 ratio (v/v). The latex collection process was designed to ensure the plants’ continued health. Subsequently, the latex was transported to the laboratory, gently agitated at 4 °C for 12 h, and centrifuged at 12,000× g at 4 °C for 15 min, resulting in a clear solution that served as the crude enzyme.
Crude LL activity in various buffer systems
The latex from C. procera demonstrated the highest lysozyme production among various latex-producing plant species, leading to its selection for investigation. Crude C. procera latex was evaluated for its activity in distinct buffer systems, each containing approximately 1.0 ml of latex per 100 mM of buffer. These buffer systems included glycine–HCl (pH 2.0, 2.5, and 3.0), citrate phosphates (pH 3.5, 4, and 4.5), sodium acetate (pH 5.0 and 5.5), sodium phosphate (pH 6.0, 6.5, and 7.0), and Tris–HCl (pH 7.5, 8.0, and 8.5). Latex was suspended in the previously described buffers and then centrifuged (12,000× g at 4 °C for 20 min) before to analyse the activity of lysozyme. Other analyses were conducted on the buffer system showing the highest activity.
Ammonium sulfate precipitation of crude LL activity
The C. procera latex was homogenized and centrifuged (12,000× g at 4 °C for 15 min); in this procedure was used a Tris–HCl buffer (pH 8.5) with a concentration of 100 mM measured at at 4 °C. Lysozyme’s notable effects were evaluated across different ammonium sulfate precipitation ranges (0–20%, 20–40%, 40–60%, 60–80%, and 80–100% w/v). It was found that the highest lysozyme activity was observed in the 60–80% range compared to other precipitation levels. After precipitation, the protein was separated by centrifugation at 12,000× g for 20 min at 4 °C and subsequently subjected to desalination using a dialysis membrane against a Tris–HCl buffer with a pH of 8.5 and a concentration of 10 mM for a period of 24 h. Protein homogeneity was assessed through SDS‒PAGE analysis. The supernatant, containing substantial protein content, was freeze-dried and reserved for further examination.
Purification of LL obtained from C. procera
CM-cellulose column chromatography
The proteins, obtained through precipitation with freeze-dried ammonium sulfate, were introduced onto a CM-Cellulose column (Catalog No. C0806; Sigma Aldrich, US; dimensions 20.0 × 2.0 cm; length × width). This column had been pre-equilibrated with Tris–HCl buffer (100 mM; pH 8.5). The proteins that adhered to the column were eluted using a gradient of NaCl ranging from 0.0 to 0.5 M. We collected 2.0 ml fractions at a flow rate of 0.250 ml per minute. Subsequently, the protein content and lysozyme activity of each fraction were assessed using a UV–Visible spectrophotometer (Hitachi, H-2900, Japan). We consolidated all the active fractions, subjected them to desalting, and freeze-dried.
Sephadex G-100 column chromatography
The active fractions obtained from the CM-Cellulose column, after desalination and freeze-drying, were introduced into a Sephadex G-100 column (Catalog No. G10050; Sigma Aldrich, USA; dimensions 80.0 × 1.5 cm; length × breadth) previously conditioned with Tris–HCl buffer (100 mM; pH 8.5). Subsequently, the proteins were eluted from the column and collection of all fractions (2.0 ml each) occurred at a flow rate of 0.3 ml per minute. The fractions exhibiting activity and uniformity were consolidated, followed by dialysis and freeze-drying. The resulting freeze-dried protein samples were preserved at − 20 °C for future use. The enzyme of high purity obtained underwent assessment for both protein content and lysozyme activity.
Protein assay
The protein concentration was evaluated at various purification stages through spectrophotometric analysis, specifically measuring absorbance at both 280 nm and 595 nm. This assessment followed the Bradford method with bovine serum albumin as a reference standard [24].
Lysozyme assay
Lysozyme activity was assessed using spectrophotometry in accordance with Jiang et al.'s method [25]. The evaluation involved measuring the rate of Micrococcus lysodeikticus suspension lysis. To prepare the assay, a known quantity of macromolecule (0.5 mg) and the specified substrate (0.3 ml) were combined, and the volume was adjusted to 3.0 ml with potassium phosphate buffer (50 mM; pH 6.5). After incubating the mixture at 37 °C for 1 h, the lysozyme activity was tracked at 570 nm using a Hitachi H-2900 spectrophotometer from Japan. Lysozyme activity was quantified as the reduction in absorbance at 570 nm per minute during the catalytic hydrolysis of the Micrococcus lysodeikticus suspension, equating to a 0.001 absorption decrease per minute.
SDS‒PAGE analysis
To evaluate the uniformity and integrity of the enzymes, we conducted SDS‒PAGE (10%) analysis following the Laemmli method under reducing conditions. The protein bands were made visible on the gel through staining using CBB R-250 [26].
Zymographic activity
The lytic effect of lysozyme on SDS-PAGE without reducing agents was elucidated as detailed in Ansari’s work [27]. The resolving gel was enriched with lyophilized Micrococcus lysodeikticus substrate at a concentration of 0.2% (w/v). Following electrophoresis, the gel underwent a 16-h incubation at 37 °C with gentle agitation in a sodium phosphate buffer (50 mM; pH 5.2) supplemented with 1.0% (v/v) Triton X-100. This resulted in the manifestation of lysozyme’s enzymatic activity within the gel, evident in the grayish-dark background [28].
2-D gel electrophoretic analysis
An investigation was conducted using 2-D gel electrophoresis to confirm whether the isolated protein existed in its monomeric state. In the initial step of protein separation, SDS-PAGE was employed. Subsequently, the gel slice containing the separated protein underwent a 10-min incubation with SDS sample buffer, excluding β-mercaptoethanol. The second phase of protein separation was executed through SDS-PAGE on a separating gel [29]. Following the separation process, the proteins were made visible through silver nitrate staining [30].
Physicochemical characteristics of purified LL
Effect of pH
To ensure the integrity of the study, we conducted experiments with the purified LL enzyme, subjecting it to a diverse range of pH levels spanning from 3.0 to 10.0. These pH adjustments were achieved using various buffers: citrate phosphate (at pH 3.0, 3.5, and 4.0), sodium acetic acid (at pH 4.5, 5.0, and 5.5), sodium phosphate (at pH 6.0, 6.5, and 7.0), Tris–HCl (at pH 7.5, 8.0, and 8.5), and glycine–NaOH (at pH 9.0, 9.5, and 10.0), all converging to a final concentration of 50 mm. Following this, the purified LL enzyme, maintained at a concentration of 10.0 µg/ml, underwent incubation with the aforementioned buffers at a temperature of 37 °C for a duration of 1 h. Subsequently, we evaluated the remaining enzyme activity of the samples after incubation using standard examination conditions.
Effect of temperature
The purified lysozyme was subjected to incubation at a range of temperatures, starting from 10 °C and increasing in increments of 10 °C, up to a maximum of 100 °C, for a duration of 1 h. The lysozyme, purified and prepared at a concentration of 10 µg/ml, was dissolved in a Tris–HCl buffer with a pH of 8.5 (100 mM) and exposed to varying temperature conditions, as previously mentioned, for a duration of 60 min. To evaluate the lysozyme activity was applied the method suggested by [25].
Effect of metallic ions
It was studied the impact of metallic ions, i.e.: calcium (Ca2+), iron (Fe2+), silver (Ag2+), mercuric (Hg2+), zinc (Zn2+), copper (Cu2+), magnesium (Mg2+), silver (Ag2+), lead (Pb2+), and manganese (Mn2+). The concentration of each was of 10 mM, on purified LL. The solutions were incubated for 1 h (37 °C) and a control solution was prepared without metallic ions.
Effect of organic solvents
The evaluation of the stability of the purified LL solution when exposed to different organic solvents (10%), including 2-propanol, acetone, benzene, dichloromethane, dimethyl sulfoxide, ethanol, ethyl acetate, hexane, methanol, and toluene, was conducted by incubating it at 37 °C for 1 h. The assessment of residual activity was carried out under optimal assay conditions, with the enzyme’'s activity in the absence of any organic solvent used as a reference point (100%).
Antibacterial characteristics of purified LL
Procurement of microbial type cultures
The effectiveness of the antibacterial properties was assessed against a range of pathogenic bacteria, encompassing Bacillus cereus, Bacillus subtilis, Staphylococcus aureus, Bacillus licheniformis, Bacillus megaterium, Staphylococcus lentus, Micrococcus luteus, Micrococcus lylae, Enterococcus faecalis, Enterococcus hirae, Escherichia coli, Pseudomonas aeruginosa, Pseudomonas syringae, Pseudomonas fluorescens, Klebsiella pneumoniae, Agrobacterium tumefaciens, Rhodococcus fascians, Rhodococcus erythropolis, Clostridium acetobutylicum, and Salmonella typhi. These bacterial strains were procured from the Microbial Type Culture Collection & Gene Bank (MTCC) based in Chandigarh, India. The cultures were propagated and preserved on nutrient agar medium with regular sub-culturing every 2 weeks.
Antibacterial efficacy of purified LL against various pathogenic bacteria
The antibacterial efficacy of the purified LL was assessed against pathogenic bacteria using the agar well diffusion technique. Pathogens were cultivated in Luria Bertani (LB) broth, reaching a cell density of 1 × 106 cfu/ml with an OD of 0.1 at 600 nm absorbance. A 10.0 µl aliquot of the culture was subsequently combined with 5.0 ml of freshly prepared LB broth and left to incubate overnight. Following this, 0.1 ml of the overnight bacterial culture was applied to Muller Hinton agar medium (MHA), and sterile conditions were maintained as 6.0 mm wells were created in the agar medium. Finally, 50 µg ml−1 of purified LL was introduced into these wells. The Petri dishes were then incubated at 37 °C for 2 days, and the presence of a clear zone of inhibition surrounding the wells and the control area was measured and documented.
Determination of MIC of purified LL
The determination of the MIC (minimum inhibitory concentration) for the purified LL was carried out through a uniform broth micro dilution assay. To achieve this, we began by cleansing overnight microorganism cultures with PBS buffer (10 mM; pH 7.5), subjecting them to centrifugation at 3000× g for 10 min at room temperature, and then re-suspending them in PBS buffer (10 mM; pH 7.5) at a cell density of 1 × 106 cells per cfu/ml for another 10 min. Next, 100 mg of the purified LL were dissolved in 1.0 ml of Tris–HCl buffer (100 mM; pH 8.5), followed by membrane filtration (0.22 μm) to ensure sterility. Subsequently, we conducted twofold serial dilutions using sterile Tris–HCl buffer (100 mM; pH 8.5). The evaluation of purified LL activity was accomplished spectrophotometrically by assessing the reduction in OD at 600 nm. The assay took place in a 96-well plate, where the reaction mixture consisted of 100 µl of bacterial culture suspension (OD 0.1) in PBS buffer (pH 7.5) and 100 µl of the purified enzyme solution. The optical density at 600 nm was determined during the incubation (24 h and 37 °C). The control sample was prepared with 100 µl of buffer instead of the LL. After the incubation time, it was employed a Multi-Scan MK13 microplate reader from Thermo Scientific, USA, to measure the 96-well plate’s absorbance at 600 nm.
Bacterial kinetics to assess antibacterial efficacy
A standardized micro-dilution test was used to evaluate the MIC of the purified LL. The method involved a liquid growth inhibition by applying the procedure proposed by Muthu [31]. Each bacterial cell suspension was adjusted to reach the final cell density of 1 × 106 cfu/ml. After purification, the LL (together with the corresponding bacterial cultures) was incubated in sterile 96-well plates containing 100 µl of the medium and 100 µl of the bacterial cultures. After that, the plates were located for 0, 1, 2, 3, 4, 5, 6, 7, and 8 h at room temperature. An equal volume of PBS buffer (10 mM; pH 7.5) was used as a control. The optical density of the cultures was measured at 600 nm using a microplate scanner, and antibacterial activity was calculated in units using the following formula: Units (U) = [AC − AS/AC] 1/2, where AS represents the sample’s absorption, and AC denotes the absorption of the control.
Isolation of bacterial cell wall
The microorganism cultures were suspended in a PBS buffer with a concentration of 10 mM and a pH of 7.5. At this point the microorganism cultures were suspended in a PBS buffer (10 mM as a concentration and 7.5 as a pH).
Following that, a solution of 3.0 N HCl in a ratio of 20:1 (volume to weight) was employed to dissolve the pellet, which was then hydrolyzed for 15 min at 80 °C. Following the hydrolysis process, the sample underwent filtration using robust glass model filters and was allowed to cool. The resulting samples were transferred into 100 ml spherical flasks, processed at a temperature of 55 °C, and stored at a temperature of − 20 °C until they were ready for further utilization.
Determination of bacterial peptidoglycan
Ten micrograms of the bacterial cell wall were combined with 10.0 µg of purified LL and subjected to incubation at ambient room temperature. Following incubation, the solution’s volume was adjusted to 0.3 ml using 1.0 M H2SO4 and hydrolyzed at 80 °C for a duration of 20 min. Subsequently, the samples were stored under refrigeration conditions, and 5.5 ml of concentrated H2SO4 was introduced into the ground glass stoppered region, vigorously shaken, and incubated at 80 °C for 10 min. The sample was then returned to room temperature, and control tests were conducted using 0.1 ml of copper sulfate solution and 0.1 ml of p-hydroxydiphenyl solution. Incubation of the samples at room temperature continued for 20 min, and their optical absorbance was measured at 560 nm utilizing a UV spectrophotometer (Hitachi, H-2900, Japan). To evaluate the bacterial peptidoglycan content, the reduction in the intensity of the blue color was observed and quantified spectrophotometrically.
HR-TEM analysis of purified LL treated with bacterial cells
The exponential growth phase of Bacillus cereus and Escherichia coli bacterial cultures was adjusted to a concentration of 1 × 106 cells per ml (cfu ml−1). Following this, 10 µg ml−1 of purified LL was introduced to 1.0 ml of the aforementioned bacterial cultures and allowed to incubate at room temperature for 1 h. Subsequently, the sample underwent centrifugation at 3000× g for 3 min, leading to the collection of the cell pellet. About 10 µl of the solution mixture, which included purified LL treated with bacteria, was applied to a copper grid coated with glow-discharged formvar. This grid was then kept at room temperature for 1 min. To serve as a control, untreated bacteria were used. Any excess liquid was carefully removed using sterile filter paper, and the specimens were air-dried for 5 min. The examination of these samples was conducted using an energy-filtering HR-TEM (Hitachi H-7000, Japan) operating at an accelerating voltage of 120 kW. Zero-loss energy-filtered images were recorded utilizing a 4 kW slow-scan charge-coupled device camera (4000 SP; Gatan, Pleasanton, CA, USA).
Statistical analysis and reproducibility
The research work involved conducting experiments in triplicate, and the reported values indicate the average of three biological replicates, unless specified otherwise. The data is presented as means with their corresponding standard deviations. We conducted statistical analysis using Student's t-test with GraphPad Prism. The HR-TEM images provided in the study are representative of a minimum of three biological replicates.
Results
Screening of crude LL from latex-producing plants
In the current study, we assessed the lysozyme activity of 14 distinct latex-producing plant varieties collected from various regions within Chennai, Tamil Nadu, India. From this array of plant species, C. procera exhibited notably high lysozyme activity, registering at 21.767 U ml−1 (as indicated in Table 1) in comparison to the other plants under scrutiny. Consequently, we selected C. procera as the focal point for our research and subsequent exploration.
Various buffer system and ammonium sulfate precipitation analyses of crude LL
The crude latex extracted from C. procera was gathered using six distinct buffers spanning a range of pH values. Following this, a lysozyme purification process was applied. Notably, the concentrate obtained under the conditions of 100 mM Tris–HCl buffers at pH 8.5 displayed notably heightened lysozyme activity at 22.874 U ml−1, surpassing the activity observed in the alternative buffer solutions (Table 2). Moreover, the protein fractionated using 60–80% ammonium sulfate demonstrated superior lysozyme activity at 100.39 U ml−1 compared to proteins obtained using other saturation levels (Table 3).
Purification of LL obtained from C. procera
CM-cellulose column chromatography
A further purification step involved using the protein fraction obtained at 60–80% saturation, and this was achieved through CM-Cellulose ion exchange column chromatography. Subsequently, we combined approximately 100 protein fractions, each of 2.0 ml volume, from the CM-Cellulose column and assessed them for lysozyme activity using the well diffusion assay. We then pooled the active protein fractions falling within the range of 79–89, underwent dialysis against Tris–HCl buffer (10 mM; pH 8.5), and finally lyophilized them. The yield percentage was determined to be 43.69%, with a purification efficiency of 7.33%. In addition, the complete lysozyme activity and specific activity were measured at 4647.24 U and 282.95 U mg−1, respectively, as indicated in Figs. 1 and 3; lane 1, and Table 4. Following this, we observed a single retention peak for the fractions, and the protein pool from the CM-Cellulose column underwent further chromatography using gel filtration column chromatography.
Sephadex G-100 column chromatography
A total of 100 protein fractions, each measuring 2.0 ml, were collected from the gel filtration column. These fractions underwent assessments for their protein content and lysozyme activity. The active fractions, specifically those falling within the range of 76–86, were amalgamated, subjected to dialysis using Tris–HCl buffer (10 mM; pH 8.5), and subsequently lyophilized. The resulting figures for yield and purification stood at 37.79% and 10.3%, respectively. In addition, the total lysozyme activity and specific activity were determined as 4019.99 U and 397.86 U mg−1, correspondingly, as illustrated in Figs. 2 and 3; lane 2 and Table 4.
Molecular mass analyses of SDS‒PAGE, zymogram and 2-D gel electrophoresis
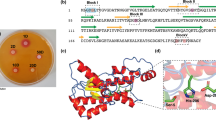
An isolated absorption peak was noted within the range of fractions 76–86, indicating the purity of a singular protein entity. In the SDS‒PAGE gel, the protein sample displayed a uniform band upon silver staining, confirming its singularity. By comparing its relative mobility to a standard molecular mass protein, the molecular weight of the purified LL protein was determined to be 14.4 kDa (Fig. 3; lane 2). The zymographic assay, conducted under non-reducing conditions on SDS‒PAGE using LL substrate, corroborated this molecular weight at 14.4 kDa (Fig. 3; lane 3), as evidenced by the distinct clear zone resulting from substrate utilization. The electrophoretic examination in a 2-D setting further affirmed the molecular mass of the purified LL to be 14.4 kDa (Fig. 4). Within physiological processes, proteins and enzymes with an acidic nature, linked to metabolism and defense, are found in a soluble state. The purified LL demonstrated an acidic nature, evident by its resolution at a pH of 5.5 on the IEF strip (Fig. 4).
Physicochemical characteristics of the purified LL
Effect of pH
The purified LL demonstrated remarkable resilience, maintaining over 83% of its initial activity across a wide spectrum of pH levels spanning from 3.0 to 10.0. Subsequently, its activity gradually declined with higher pH values, ultimately reaching its nadir at pH 10.0, where it exhibited only 30% of its relative activity. These findings unmistakably highlight the acidic lysozyme’s stability within the pH range of 4.6–6.5, while revealing its diminished stability within the pH range of 8.5–10.0, as depicted in Fig. 5a.
Effect of temperatures
Figure 5b depicts the impact of varying temperatures on the purified LL, showing a gradual rise in relative activity as the temperature increases from 10 to 50 °C, followed by a decline in lysozyme activity at its lowest point of 100 °C, as indicated in Fig. 5b.
Effect of metallic ions
The presence of ions like Mg2+ and Ca2+ led to a mild reduction in the enzyme’s relative activity, reducing it to around 83.58% and 63.88%, respectively, as shown in Fig. 5c. In contrast, Hg2+ significantly diminished the LL enzyme’s activity when present at a concentration of 10 mM, preserving only a maximum of 25.41% of its relative enzymatic function. This observation indicates the distinct effects of these ions on the enzyme’s performance.
Effect of organic solvents
The outcomes depicted in Fig. 5d exhibit the impact of various organic solvents on the activity of purified LL. The stability in organic solvents was observed through exposure to 2-propanol, dimethyl sulfoxide, acetone, ethanol, toluene, dichloromethane, methanol, ethyl acetate, benzene, and hexane, displaying corresponding activities of 86.82%, 76.18%, 65.35%, 57.73%, 56.24%, 49.57%, 41.24%, 35.88%, 26.72%, and 15.27% respectively (as illustrated in Fig. 5d). Notably, treatment with dichloromethane, methanol, ethyl acetate, benzene, and hexane significantly diminished the enzyme activity of the purified protein compared to the control (Fig. 5d).
Antibacterial studies of purified LL against pathogenic bacteria
Antibacterial efficacyof purified LL against various pathogenic bacteria
The results presented in Table 5 demonstrate that the antibacterial efficacy of the purified LL exhibited zone of inhibition measurements ranging from 15 to 24 mm against various pathogenic bacteria, including Bacillus cereus, Bacillus subtilis, Bacillus megaterium, Bacillus licheniformis, Staphylococcus aureus, Staphylococcus lentus, Micrococcus luteus, Micrococcus lylae, Enterococcus faecalis, Enterococcus hirae, Escherichia coli, Pseudomonas aeruginosa, Pseudomonas syringae, Pseudomonas fluorescens, Klebsiella pneumoniae, Agrobacterium tumefaciens, Rhodococcusfascians, Rhodococcu serythropolis, Clostridium acetobutylicum, and Salmonella typhi, respectively.
MIC determination of purified LL
The MIC for the purified LL was tested against 20 different pathogenic bacteria, including B. cereus, B. subtilis, B. megaterium, B. licheniformis, S. aureus, S. lentus, M. luteus, M. lylae, E. faecalis, E. hirae, E. coli, P. aeruginosa, P. syringae, P. fluorescens, K. pneumoniae, A. tumefaciens, R. fascians, R. erythropolis, C. acetobutylicum, and S. typhi. The MIC values ranged from 13 to 30 µg ml−1, with specific values of 13, 15, 18, 23, 25, 22, 18, 23, 25, 22, 14, 17, 26, 28, 30, 29, 22, 24, 26, and 25 µg ml−1, respectively (Table 5).
Bacterial kinetics of purified LL on antibacterial activity
The research investigated the impact of purified LL derived from C. procera on the antimicrobial efficacy against 20 distinct pathogenic bacteria. Within a 3-h incubation period with the purified LL, the growth of all bacterial strains experienced a consistent decline, and extended incubation exceeding 3 h led to the complete suppression of bacterial growth. Moreover, we quantified and expressed the antibacterial activity in terms of units relative to the incubation duration. Notably, the most significant reduction in bacterial cell counts was observed after 8 h, with purified LL displaying 1.894 units for B. cereus, 1.667 units for B. subtilis, 1.839 units for E. coli, and 1.463 units for P. aeruginosa (Fig. 6).
Determination of peptidoglycan
The peptidoglycan from the bacterial cell wall was extracted and exposed to purified LL at a concentration of 10.0 μg ml−1. Subsequent spectrophotometric analysis revealed significant degradation of the peptidoglycan. The degradation percentages were as follows: 66.67%, 64.16%, 61.58%, 56.48%, 53.89%, 53.91%, 43.68%, 43.68%, 43.66%, 41.11%, 41.09%, 38.56%, 35.99%, 35.99%, 35.97%, 33.44%, 33.44%, 28.32%, 25.76%, and 23.18% for respective bacterial strains (B. cereus, E. coli, B. subtilis, P. aeruginosa, B. megaterium, M. luteus, E. hirae, R. fascians, S. lentus, M. lylae, B. licheniformis, R. erythropolis, E. faecalis, S.typhi, S. aureus, C. acetobutylicum, P. syringae, P. fluorescens, A. tumefaciens, and K. pneumoniae). A summarized overview of the purified LL’s impact on bacterial peptidoglycan can be found in Table 5.
HR-TEM analysis of purified LL treated with bacterial cells
The purified LL caused significant damage to the bacterial cell membranes of both B.subtilis and E. coli. This damage was evident as the membranes appeared shrunken, and the cytoplasmic contents leaked out, adhering to the cell surface (refer to Fig. 7b and d). In contrast, the control culture displayed smooth and structurally intact cell surfaces (see Fig. 7a and c). Bacterial cells treated with the purified LL exhibited corrugated surfaces, marked membrane disruptions, and deep craters. Importantly, leakage of cytoplasmic and nuclear contents was observed at these disrupted sites. In contrast, the membranes of control cells remained undamaged, with nuclei intact. Therefore, this study provides unequivocal evidence of the purified LL’s ability to disrupt bacterial cell membranes (see Fig. 7a–d).
Discussion
Latex-producing plants can be found across the globe in various plant families, including Apocynaceae, Asteraceae, Caricaceae, Euphorbiaceae, Moraceae, and Papaveraceae. These plants possess specialized lactiferous structures. Latex, which is commonly known as aqueous suspensions or emulsions, has a diverse composition that includes alkaloids, tannins, oils, terpenoid compounds like resins and rubber, starches, sugars, and proteins [32]. Extensive research has delved into the exploration of hydrolytically active proteins, such as proteases, lysozyme, and chitinase, which are produced by plants as a defense mechanism [33,34,35]. In this current investigation, we endeavored to isolate, purify, and characterize the LL from the latex of C. procera. This LL, demonstrating significant antibacterial efficacy against a diverse range of pathogenic bacteria, was subject to molecular and physicochemical analysis. The quest for natural, eco-friendly antibiotic agents from nature has surged in recent times due to the high costs of synthetic antibiotics and the escalating bacterial resistance to these pharmaceuticals [36,37,38].
In pursuit of the aforementioned objective, a research inquiry was launched, involving the extraction of crude latex from 14 distinct latex-producing plant species and the assessment of lysozyme activity. Among the latex extracts derived from latex-producing plant species that exhibited lysozyme activity, the latex extracted from C. procera demonstrated comparatively elevated lysozyme activity (21.767 U ml−1; as detailed in Table 1). A previous study conducted by Sytwala et al. documented the investigation of lysozyme activity in the latex of 110 different species belonging to the Euphorbia genus [32]. The distinctive attributes of LL primarily hinge on the diverse physicochemical characteristics of latices. The presence of ionic constituents in the buffer can impact the discharge of bioactive substances by altering the enzyme’s stability and solubility. Evaluating if the constituents in the buffer affect the lysozyme functionality of C. procera, the latex extracts obtained were subjected to various buffer solutions spanning pH levels from 2.0 to 10.5 for analysis. As Table 2 clearly indicates, the utilization of Tris–HCl buffer (100 mM; pH 7.5) resulted in a notably elevated lysozyme activity of 22.874 U ml−1. This increase in lysozyme activity can be attributed to the enhanced solubilization and increased solubility of the protein content (1.0 mg ml−1) facilitated by the buffering system. Quan et al.'s research findings also support the notion that lysozyme exhibits considerably higher activity when exposed to Tris–HCl buffer [39], and Kumaran Subramanian et al. have similarly demonstrated the efficacy of Tris–HCl buffer in boosting lysozyme activity [40]. Efforts have been exerted in the current study to purified LL using established protein purification methods and cutting-edge techniques. Initially, the crude latex sourced from C. procera was subjected to ammonium sulfate fractionation at varying saturation levels as a first step in the purification procedure. It was observed that proteins obtained at 60–80% saturation exhibited notably elevated lysozyme activity (100.39 U ml−1; as indicated in Table 3). The relationship between the saturation level of the precipitant and the protein's molecular mass was notable, showcasing that higher saturation led to the precipitation of proteins with higher molecular masses, while lower saturation favored proteins with lower molecular masses [41, 42].
The protein fraction acquired through 60–80% saturation was employed for further purification using CM-Cellulose and gel filtration column chromatography. The active fractions clearly indicate the presence of the lysozyme enzyme in the latex of C. procera. In addition, these active fractions underwent analysis through SDS‒PAGE. As depicted in Fig. 3, the lysozyme from C. procera latex displayed a band (lane 1, larger band) at approximately 14.4 kDa. However, to achieve lysozyme devoid of contaminants, it was necessary to remove the small quantity of observed contaminating proteins. Therefore, an extra purification stage proved essential, and the combined active fractions acquired from the preceding purification phase were employed to conduct additional purification via gel filtration column chromatography. The lysozyme isolated through the gel filtration column exhibited a distinct, uniform band (as depicted in Figs. 2 and 3; Lane 2) on SDS‒PAGE, resulting in a final yield of 37.79% (refer to Table 4). The efficiency of the CM-Cellulose column chromatography method and the gel filtration column chromatography method was found to be consistent with the purification of lysozyme from rubber latex allergens, as outlined in reference [43]. The molecular weight of the purified LL was determined to be 14.4 kDa through SDS‒PAGE analysis, where we estimated its mobility value (Rf value) in comparison to standard molecular mass proteins (as shown in Fig. 3). This molecular weight for purified LL closely resembled lysozyme, which has a known molecular weight of 27 kDa when sourced from rubber latex allergens [44, 45], Heveabrasiliensis latex (26 kDa) [20], and AsclepiasSyria ca latex (28 kDa) [23]. Furthermore, this consistency in molecular weight was corroborated by the zymographic assay conducted with hen egg-white lysozyme (14.4 kDa) [46, 47]. Therefore, it can be concluded that LL exhibits molecular weights ranging from 14 to 28 kDa.
The remarkable resilience of acidic lysozyme in the alkaline pH range is in stark contrast to how lysozyme typically behaves in the majority of plant species [48]. LL demonstrated consistent activity between pH 4.6 and 6.5, but it failed to exhibit lysozyme activity when subjected to pH levels below 3.0 or above 9.0, potentially attributable to an incorrect protonation state in the amino acid side chains during enzyme activation. This elucidates the necessity for an acidic environment to achieve its optimal lysozyme performance (see Fig. 5a). Our findings align with the research of Yanan Wang and Nasim Khorshidian [49, 50] as well as Manikandan [51], which highlighted that lysozyme extracted from Momordica charantia fruits displayed its peak lysozyme activity in the pH range spanning from 5.5 to 5.8. Temperature significantly influences the biological activity of enzymes and proteins. The purified LL demonstrated its highest enzymatic activity when subjected to an incubation temperature of 50 °C, but gradually lost its capacity to perform enzymatic functions when exposed to temperatures exceeding 65 °C for 1 h, as depicted in Fig. 5b. This temperature profile of the purified LL closely resembled that of the antimicrobial protein extracted from Clitocybe sinopica, wherein the protein became inactive after just 30-min of incubation at 80 °C [52]. However, it's worth noting that the heat resistance of the purified LL was remarkably comparable to that of plant lysozyme obtained from Momordica charantia L, as documented by Wang [53]. Metallic ions are widely recognized for their capacity to impart stability to enzymes and are proposed to offer defense against protein autolysis. Nevertheless, our investigation revealed that these metal ions hindered C. procera LL’s function, with the exception of Mg2+, which sustained the activity of purified LL, as depicted in Fig. 5c. These findings align with those of Pengfei Zhang et al., who reported that Mg2+ had a modestly enhancing effect on lysozyme activity sourced from the eastern oyster’s plasma. Treating the purified enzyme with organic solvents like benzene and hexane led to a complete loss of enzyme activity, as demonstrated in Fig. 5d [54]. In general, the presence of organic solvents can at times diminish the enzyme’'s structural flexibility by disrupting covalent interactions, which are vital for catalytic processes. This can be attributed to the deterioration of structural integrity and destabilization of the protein’s polar transition state [55].
The MIC values for the purified lysozyme found in C. procera latex were 13 µg ml−1 against B. cereus (a gram-positive bacterium) and 14 µg ml−1 against E. coli (a gram-negative bacterium). These MIC values were comparable to those of lysozyme derived from cauliflower, which had been tested against various harmful bacteria [51]. Lysozyme extracted from plant sources has demonstrated significant antibacterial effects against a wide spectrum of both gram-positive and gram-negative bacteria [49, 56], and it has found extensive applications in the agricultural and industrial sectors [57, 58]. We investigated the antibacterial activity kinetics of the purified lysozyme against 20 different pathogenic bacteria. Among these, the most substantial reduction in bacterial cell counts was observed in B. cereus, B. subtilis, E. coli, and P.aeruginosa, with reductions of 1.894 units at 8 h, 1.667 units at 8 h, 1.839 units at 8 h, and 1.463 units at 8 h, respectively (as shown in Fig. 6a and b). The kinetics of antibacterial activity were assessed following the method outlined by Muthu et al. [29, 31].
The purified LL effectively lysed the peptidoglycan in the cell walls of 20 different pathogenic bacteria, as demonstrated in Table 5. Among these, B. cereus exhibited the highest peptidoglycan degradation at 66.67%, while K. pneumoniae displayed the least degradation at 23.18%. In contrast, untreated cells maintained their normal bacterial surface architecture. Our HR-TEM electron microscopy study provided clear evidence of the interaction between the purified LL and the bacterial cell wall, leading to the release of cytoplasmic content into the surrounding medium. In addition, a reduction in peptidoglycan content in the bacterial cell wall indicated that LL had disrupted and destabilized the cell membrane, ultimately causing cell death, as shown in Fig. 7b and d. A similar impact on cell architecture was observed when lysozyme was used against Pseudomonasaeruginosa, Acinetobacterbaumannii, and methicillin-resistant Staphylococcusaureus, as previously reported [59, 60].
Conclusion
In conclusion, we present a plant-derived lysozyme obtained from the latex of C. procera, exhibiting robust antibacterial properties against 20 distinct pathogenic bacteria. The purified lysozyme exists as a monomeric form with a molecular mass of 14.4 kDa. The stability of the purified lysozyme was evaluated across different buffer solutions, metallic ions, temperature ranges, and organic solvents. Furthermore, the HR-TEM image clearly demonstrates the effective inhibitory impact of the lysozyme on pathogenic bacteria, resulting in noticeable cell content leakage. The remarkable bactericidal activity underscores the potential applications of this latex-derived lysozyme in various sectors, including agriculture, pharmaceuticals, and biotechnology.
Data availability
The authors declare availability of data and material.
References
Hua J, Liu Y, Xiao CJ, Jing SX, Luo SH, Li SH (2017) Phytochemistry 136:56
Gracz-bernaciak J, Mazur O, Nawrot R (2021) Int J Mol Sci 22:12427
Salomé Abarca LF, Klinkhamer PGL, Choi YH (2019) Planta Med 85:856
Agrawal AA, Hastings AP (2019) J Chem Ecol 45:1004
Ozber N, Carr SC, Morris JS, Liang S, Watkins JL, Caldo KM, Hagel JM, Ng KKS, Facchini PJ (2022) Nat Commun 13:1
Konno K (2011) Phytochemistry 72:1510
Benninghaus VA, Van Deenen N, Müller B, Roelfs KU, Lassowskat I, Finkemeier I, Prüfer D, Gronover CS (2020) J Exp Bot 71:1278
Sinha M, Singh RP, Kushwaha GS, Iqbal N, Singh A, Kaushik S, Kaur P, Sharma S, Singh TP (2014) Sci World J 2014:1
Ramos MV, Demarco D, da Costa Souza IC, de Freitas CDT (2019) Trends Plant Sci 24:553
Kaur A, Batish DR, Kaur S, Chauhan BS (2021). Front Plant Sci. https://doi.org/10.3389/fpls.2021.690806
Al Sulaibi MAM, Thiemann C, Thiemann T (2020) Open Chem J 7:1
Shobowale OO, Ogbulie NJ, Itoandon EE, Oresegun MO, Olatope SOA (2013) Phytochemical and antimicrobial evaluation of aqueous and organic extracts of Calotropis procera ait leaf and latex. Niger Food J 31(1):77–82
Midoro-Horiuti T, Brooks EG, Goldblum RM (2001) Ann Allergy Asthma Immunol 87:261
Tan SY, Tatsumura Y (2015) Singap Med J 56:366
Coutinho HDM, Lôbo KM, Bezerra DAC, Lôbo I (2008) Indian J Pharmacol 40:3
Nawaz N, Wen S, Wang F, Nawaz S, Raza J, Iftikhar M, Usman M (2022) Lysozyme and its application as antibacterial agent in food industry. Mol 27(19):6305
Li J, Hu S, Jian W, Xie C, Yang X (2021). Bot Stud. https://doi.org/10.1186/s40529-021-00312-x
Broekaert WF, Cammue BPA, De Bolle MFC, Thevissen K, De Samblanx GW, Osborn RW, Nielson K (1997) CRC Crit Rev Plant Sci 16:297
Kanokwiroon K, Teanpaisan R, Wititsuwannakul D, Hooper AB, Wititsuwannakul R (2008) Mycoses 51:301
Martin MN (1991) Plant Physiol 95:469
Islam A, Ma AM, Parvin S, Meh S, Zaman MK, Parvin F, Zaman S, Salah Uddin M (2015) Asian J Pharm Clin Res 8:308
James Brockbank W, Lynn KR (1979) BBA Protein Struct 578:13
Lynn KR (1989) Phytochemistry 28:1345
Bradford MM (1976) Anal Biochem 72:248
Jiang MF, Hu MJ, Ren HH, Wang L (2015) Asian Australas J Anim Sci 28:1774
Laemmli UK (1970) Nature 227:680
Ansari M, Zubair S, Atif S, Kashif M, Khan N, Rehan M, Anwar T, Iqbal A, Owais M (2009) Protein Pept Lett 17:11
Sakthivel M, Karthikeyan N, Palani P (2010) Detection and analysis of lysozyme activity in some tuberous plants and Calotropis procera’s latex. J Phytology 2(11):65–72
Muthu S, Gopal VB, Soundararajan S, Nattarayan K, Narayan KS, Lakshmikanthan M, Malairaj S, Perumal P (2017) Plant Physiol Biochem 112:161
Nesterenko MV, Tilley M, Upton SJ (1994) J Biochem Biophys Methods 28:239
Muthu S, Gopal VB, Karthik NS, Sivaji P, Malairaj S, Lakshmikanthan M, Subramani N, Perumal P (2017) Int J Biol Macromol 103:878
Sytwala S, Günther F, Melzig MF (2015) Plant Physiol Biochem 95:35
dos Santos C, Franco OL (2023) Plants 12:1
Sueldo DJ, Godson A, Kaschani F, Krahn D, Kessenbrock T, Buscaill P, Schofield CJ, Kaiser M, Van Der Hoorn N (2024) Trondheim. New Phytol 241:397–408
Punja ZK, Zhang YY (1993) J Nematol 25:526
Liliana S (2020) Water 12:3313
Davies J (1996) Microbiologia 12:9
Heinekamp T, Thywißen A, Macheleidt J, Keller S, Valiante V, Brakhage AA (2012) Front Microbiol 3:440
Quan L, Wei D, Jiang X, Liu Y, Li Z, Li N, Li K, Liu F, Lai L (2008) Anal Biochem 378:144
Subramanian K, Balaraman D, Panangal M, Nageswara Rao T, Perumal E, Amutha R, Kumarappan A, Sampath Renuga P, Arumugam S, Thirunavukkarasu R, Aruni W, Yousef Alomar S (2022) Sci Rep 12:1
Wingfield P (2001) Curr Protoc Protein Sci
Kramer RM, Shende VR, Motl N, Pace CN, Scholtz JM (2012) Biophys J 102:1907
Yagami T, Sato M, Nakamura A, Shono M (1995) J Allergy Clin Immunol 96:677
Kahn SL, Podjasek JO, Dimitropoulos VA, Brown CW (2016) Dis Mon 62:5
Seng YY, Yun CT, Abdullah N, Wong FWF (2022) Ind Crops Prod 177:1
Hardt M, Guo Y, Henderson G, Laine RA (2003) Anal Biochem 312:73
Callewaert L, Vanderkelen L, Deckers D, Aertsen A, Robben J, Michiels CW (2008) Appl Environ Microbiol 74:4978
Meyer K, Hahnel E, Steinberg A (1946) J Biol Chem 163:733
Khorshidian N, Khanniri E, Koushki MR, Sohrabvandi S, Yousefi M (2022). Front Nutr. https://doi.org/10.3389/fnut.2022.833618
Chatake T, Tanaka I, Kusaka K, Fujiwara S (2022) Acta Crystallogr Sect D Struct Biol 78:770
Manikandan M, Balasubramaniam R, Chun SC (2015) Appl Biochem Biotechnol 177:556
Zheng S, Liu Q, Zhang G, Wang H, Ng TB (2010) Acta Biochim Pol 57:43
Wang S, Shao B, Chang J, Rao P (2011) Eur Food Res Technol 232:613
Zhang P, He J, Zou X, Zhong Y, Pan X, Zhang J, Pang H (2022) J Environ Manag 315:1
Zhu F, He B, Gu F, Deng H, Chen C, Wang W, Chen N (2020) J Biotechnol 309:68
Ibrahim HR, Higashiguchi S, Juneja LR, Kim M, Yamamoto T (1996) J Agric Food Chem 44:1416
Cegielska-Radziejewska R, Lesnierowski G, Kijowski J (2008) Polish J Food Nutr Sci
Alhazmi A, Stevenson JW, Amartey S, Qin W (2014) Int J Biol
Saito H, Sakakibara Y, Sakata A, Kurashige R, Murakami D, Kageshima H, Saito A, Miyazaki Y (2019) PLoS ONE 14:1
Kim S, Lee DW, Jin JS, Kim J (2020) J Glob Antimicrob Resist 22:32
Funding
Open access funding provided by Università degli Studi Mediterranea di Reggio Calabria within the CRUI-CARE Agreement.
Author information
Authors and Affiliations
Contributions
For research articles with several authors, a short paragraph specifying their individual contributions must be provided. The following statements should be used “Conceptualization, SM and AMG; methodology, VG; software, AA and AMG; validation, PP and ML; formal analysis, KK; investigation, MAAN; resources, LG and AMG; data curation, SM; writing—original draft preparation, SM; writing—review and editing, LG, AA, and AMG; visualization, VG and AMG; supervision, LG; project administration, SM; funding acquisition, SM and AMG. All authors have read and agreed to the published version of the manuscript.”
Corresponding authors
Ethics declarations
Conflict of interest
The authors unanimously assert that they have no conflicts of interest.
Compliance with ethics requirements
This article does not contain any studies with human or animal subjects.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Muthu, S., Gopal, V., AL-Younis, Z.K. et al. Extraction, purification, and investigation of the antibacterial potential of lysozyme isolated from the latex of Calotropis procera. Eur Food Res Technol 250, 1401–1416 (2024). https://doi.org/10.1007/s00217-024-04467-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00217-024-04467-2