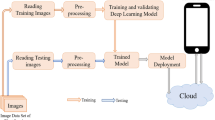
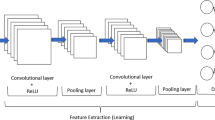
Abstract
Walnuts are widely used, although they come in a variety of types and qualities. It is essential to choose the correct walnut variety with the necessary ecological characteristics to continue the production of walnut fruit, which has positive benefits on human health. Because planting a walnut garden is expensive and the harvesting process takes a while. However, since the colour and feel of walnut leaves are so similar, it can be challenging to tell them apart. Experts must devote a significant amount of time to differentiating walnut kinds, and morphological tests should be conducted. There are different studies in the literature for walnut variety differentiation. Nevertheless, those are studies conducted with the classification of a small number of walnut varieties or laboratory experiments. With the advancement of technology, deep learning techniques based on computers are now routinely utilized for leaf recognition. These technologies enable significant reductions in error rates, time saves, and cost. With a total of 1751 leaf pictures collected from 18 species of walnuts, a special walnut dataset was constructed for this study in order to identify walnut types from walnut leaves. To automatically classify the provided dataset, images are trained with residual block-based convolutional neural network architectures. Following the discovery of each image's deep features, the Atom Search Optimization algorithm was used to choose the most distinctive characteristics. Support vector machines (SVM) were used to classify walnut species with the new feature set created. The experimental studies of the proposed model based on Residual block and Atom Search optimization successfully categorised the walnut dataset with an accuracy rating of 87.42%.







Similar content being viewed by others
References
Kulkarni K, Zhang Z, Chang L, Yang J, da Fonseca PCA, Barford D (2013) Building a pseudo-atomic model of the anaphase-promoting complex. Acta Crystallogr Sect D Biol Crystallogr 69(11):2236–2243
T. KARADENİZ, “Ordu Yöresinde yetiştirilen ceviz genotiplerinin (Juglans regia L.) seleksiyonu,” Ordu Üniversitesi Bilim ve Teknol. Derg., vol. 1, no. 1, pp. 65–74, 2011.
Khalesi S, Mahmoudi A, Hosainpour A, Alipour A (2012) Detection of walnut varieties using impact acoustics and artificial neural networks (ANNs). Mod Appl Sci 6(1):43
Esteki M et al (2017) Classification and authentication of Iranian walnuts according to their geographical origin based on gas chromatographic fatty acid fingerprint analysis using pattern recognition methods. Chemom Intell Lab Syst 171:251–258
L. U. Jun, X. Z. Han, and K. J. Wang, “Classification of Collection Walnut Based on GLCM and SVM,” in 2nd International Conference on Test, Measurement and Computational Method (TMCM 2017), 2017, pp. 276–281.
Khan MA et al (2021) Machine learning-based detection and classification of walnut fungi diseases. Intell Autom SOFT Comput 30(3):771–785
Anagnostis A et al (2021) A deep learning approach for anthracnose infected trees classification in walnut orchards. Comput Electron Agric 182:105998
Anagnostis A, Asiminari G, Papageorgiou E, Bochtis D (2020) A convolutional neural networks based method for anthracnose infected walnut tree leaves identification. Appl Sci 10(2):469
Beikmohammadi A, Faez K, Motallebi A (2022) SWP-LeafNET: a novel multistage approach for plant leaf identification based on deep CNN. Expert Syst Appl 202:117470
Dobrescu A, Giuffrida MV, Tsaftaris SA (2020) Doing more with less: a multitask deep learning approach in plant phenotyping. Front Plant Sci 11:141
Kamilaris A, Prenafeta-Boldú FX (2018) Deep learning in agriculture: a survey. Comput Electron Agric 147:70–90
Firildak K, Talu MF (2019) Evrişimsel sinir ağlarında kullanılan transfer öğrenme yaklaşımlarının incelenmesi. Comput Sci 4(2):88–95
K. He, X. Zhang, S. Ren, and J. Sun, “Deep residual learning for image recognition,” in Proceedings of the IEEE conference on computer vision and pattern recognition, 2016, pp. 770–778.
Al-Haija QA, Smadi MA, Zein-Sabatto S (2020) “Multi-class weather classification using ResNet-18 CNN for autonomous IoT and CPS applications.” Int Conf Comput Sci Comput Intell (CSCI) 2020:1586–1591
Dong Y, Zhang H, Wang C, Wang Y (2019) Fine-grained ship classification based on deep residual learning for high-resolution SAR images. Remote Sens Lett 10(11):1095–1104
Erdal E, Kayri M, Ekinci S (2019) Kısıtlı optimizasyon problemlerinin çözümü için atom arama optimizasyon algoritması. Dicle Üniversitesi Mühendislik Fakültesi Mühendislik Derg 10(3):841–851
Zhao W, Wang L, Zhang Z (2019) A novel atom search optimization for dispersion coefficient estimation in groundwater. Futur Gener Comput Syst 91:601–610
Özbay FA, Özbay E Diyabetik Retinopati Tespiti İçin Atom Arama Optimizasyonu İle Özellik Seçimi Yöntemi. Adıyaman Üniversitesi Mühendislik Bilim. Derg., vol. 9, no. 16, pp. 88–104.
ÖZDET B, Semra İ, “Akciğer Bilgisayarlı Tomografi Görüntülerinde Görüntü İşleme Uygulamaları İle Tümörlerinin Tespit Edilmesi,” Uludağ Üniversitesi Mühendislik Fakültesi Derg., vol. 27, no. 1, pp. 135–150.
Çelik Y, Başaran E, Dilay Y Identification of durum wheat grains by using hybrid convolution neural network and deep features, Signal, Image Video Process pp. 1–8, 2022.
Başaran E Classification of white blood cells with SVM by selecting SqueezeNet and LIME properties by mRMR method, Signal, Image Video Process., pp. 1–9, 2022.
Yang B, Gong R, Wang L, Yang S Support vector machine in image recognition of nursing methods for critically ill blood purification, Microprocess. Microsyst., p. 103398, 2020.
Japkowicz N, Shah M Evaluating learning algorithms: a classification perspective. Cambridge University Press, 2011.
Toğaçar M, Ergen B, Sertkaya ME, Zatürre Hastalığının Derin Öğrenme Modeli ile Tespiti. Firat Univ. J. Eng., vol. 31, no. 1, 2019.
Sokolova M, Lapalme G (2009) A systematic analysis of performance measures for classification tasks. Inf Process Manag 45(4):427–437
Moreno-Torres JG, Sáez JA, Herrera F (2012) Study on the impact of partition-induced dataset shift on $ k $-fold cross-validation. IEEE Trans Neural Networks Learn Syst 23(8):1304–1312
Refaeilzadeh P, Tang L, Liu H (2009) Cross-validation. Encycl database Syst 5:532–538
Zhang Y et al. MFCIS: an automatic leaf-based identification pipeline for plant cultivars using deep learning and persistent homology, Hortic. Res., vol. 8, 2021.
Liu YH (2018) Feature extraction and image recognition with convolutional neural networks. J Phys 1087(6):62032
Wei Tan J, Chang S-W, Abdul-Kareem S, Yap HJ, Yong K-T (2018) Deep learning for plant species classification using leaf vein morphometric, IEEE/ACM Trans. Comput. Biol. Bioinforma. 17(1): 82–90.
Szegedy C, Ioffe S, Vanhoucke V, Alemi AA (2017) Inception-v4, inception-resnet and the impact of residual connections on learning.
Khaire UM, Dhanalakshmi R (2019) Stability of feature selection algorithm: a review. J King Saud Univ Inf Sci.
Hammadi WQ, Qasim OS (2022) Hybrid binary atom search optimization approaches with statistical dependence for feature selection. Int Conf Comput Sci Softw Eng (CSASE) 2022:218–223
Zhao W, Wang L, Zhang Z (2019) Atom search optimization and its application to solve a hydrogeologic parameter estimation problem. Knowledge-Based Syst 163:283–304
Acknowledgements
In this study, we would like to thank the Institute Director Dr. Yılmaz BOZ and the staff of the Institute for allowing us to take the walnut leaf images from the Yalova Atatürk Horticultural Central Research Institute application garden. We would also like to thank Prof. Dr. Turan KARADENİZ and Dr. Tuba BAK for their help in determining the leaf types and taking the images.
Funding
This study was not funded.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there is no conflict of interest.
Ethical standards
This article does not contain any studies with human or animal subjects.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Karadeniz, A.T., Çelik, Y. & Başaran, E. Classification of walnut varieties obtained from walnut leaf images by the recommended residual block based CNN model. Eur Food Res Technol 249, 727–738 (2023). https://doi.org/10.1007/s00217-022-04168-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00217-022-04168-8