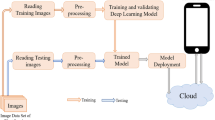
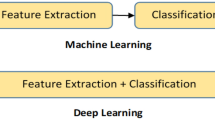
Abstract
There are many types of haricot beans, the nutrient consumed all over the world. Each type differs in terms of features such as taste, size, economic value, etc. But even if they are different types, bean grains are frequently confused with each other. For these reasons, it is important to separate the bean grains of different species. For this purpose, a haricot bean dataset consisting of 33,064 images of 14 different bean types was created. By using these images, 3 different pre-trained Convolutional Neural Networks (CNN) were trained via the transfer learning method. Within the scope of the study, InceptionV3, VGG16, and VGG19 CNN models were used. These models were utilized for both end-to-end classification and extraction of image features. Firstly, the images were classified via Inception V3, VGG16, and VGG19 models. As a result of this classification, 84.48%, 80.63%, and 81.03% classification success were obtained from InceptionV3, VGG16, and VGG19 models, respectively. Secondly, the image features of these 3 models were taken from the layer just before the classification layer. Then, these features were given as input to the Support Vector Machine (SVM) and Logistic Regression (LR) models. Images were classified using six different models, InceptionV3 + SVM, VGG16 + SVM, VGG1 + SVM and InceptionV3 + LR, VGG16 + LR, VGG1 + LR. Classification successes obtained from InceptionV3 + SVM, VGG16 + SVM, and VGG19 + SVM were 79.60%, 81.97%, 80.64%, respectively. And, the classification successes obtained from InceptionV3 + LR, VGG16 + LR, and VGG19 + LR were 82.35%, 83.71%, and 83.54%, respectively. The InceptionV3, among all models, was determined to be the best classification model with a classification success of 84.48%. On the other hand, the model with the lowest classification success was determined to be the InceptionV3 + SVM. Detailed analysis of the created models was also carried out with precision, recall, and F-1 score metrics. It is thought that the proposed models can be used to distinguish haricot bean types in a quick and accurate way. Furthermore, the proposed computer vision methods can be combined with robotic systems and used to the distinction of bean types. By means of image processing, varieties can be determined on conveyor belts, and dry bean varieties can be purified with delta robots.










Similar content being viewed by others
Data availability
The dataset used in the study can be accessed from the link https://www.muratkoklu.com/datasets/Dry_Bean_Image_Dataset.zip.
References
Long Y, Bassett A, Cichy K, Thompson A, Morris D (2019) Bean split ratio for dry bean canning quality and variety analysis. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops
Suárez-Martínez SE, Ferriz-Martínez RA, Campos-Vega R, Elton-Puente JE, de la Torre Carbot K, García-Gasca T (2016) Bean seeds: leading nutraceutical source for human health. CyTA J Food 14(1):131–137. https://doi.org/10.1080/19476337.2015.1063548
Alban N, Laurent B, Martin Y, Ousman B (2014) Quality inspection of bag packaging red beans (Phaseolus vulgaris) using fuzzy clustering algorithm. J Adv Math Computer Sci. https://doi.org/10.9734/BJMCS/2014/12981
Saha D, Annamalai M (2021) Machine learning techniques for analysis of hyperspectral images to determine quality of food products: a review. Curr Res Food Sci. https://doi.org/10.1016/j.crfs.2021.01.002
Sun J, Jiang S, Mao H, Wu X, Li Q (2016) Classification of black beans using visible and near infrared hyperspectral imaging. Int J Food Prop 19(8):1687–1695. https://doi.org/10.1080/10942912.2015.1055760
Ropelewska E, Sabanci K, Aslan MF, Azizi A (2022) A novel approach to the authentication of apricot seed cultivars using innovative models based on image texture parameters. Horticulturae 8(5):431
Janiesch C, Zschech P, Heinrich K (2021) Machine learning and deep learning. Electron Mark 31(3):685–695
Chauhan NK, Singh K (2018) A review on conventional machine learning vs deep learning. In 2018 International Conference on Computing, Power and Communication Technologies (GUCON). IEEE https://doi.org/10.1109/GUCON.2018.8675097
LeCun Y, Bengio Y, Hinton G (2015) Deep learning. Nature 521(7553):436–444. https://doi.org/10.1038/nature14539
Shahin M, Symons S (2003) Lentil type identification using machine vision. Canad Biosyst Eng 45:3.5
Ghamari S (2012) Classification of chickpea seeds using supervised and unsupervised artificial neural networks. Afr J Agric Res 7(21):3193–3201. https://doi.org/10.5897/AJAR11.2071
Kumar M, Bora G, Lin D (2013) Image processing technique to estimate geometric parameters and volume of selected dry beans. J Food Measur Character 7(2):81–89. https://doi.org/10.1007/s11694-013-9142-7
Daskalov P, Kirilova E, Georgieva T (2018) Performance of an automatic inspection system for classification of Fusarium Moniliforme damaged corn seeds by image analysis. In MATEC Web of Conferences. EDP Sci https://doi.org/10.1051/matecconf/201821002014
Wu A, Zhu J, Yang Y, Liu X, Wang X, Wang L, Zhang H, Chen J (2018) Classification of corn kernels grades using image analysis and support vector machine. Adv Mech Eng 10(12):1687814018817642. https://doi.org/10.1177/1687814018817642
Li X, Dai B, Sun H, Li W (2019) Corn classification system based on computer vision. Symmetry 11(4):591. https://doi.org/10.3390/sym11040591
Pourdarbani R, Sabzi S, García-Amicis VM, García-Mateos G, Molina-Martínez JM, Ruiz-Canales A (2019) Automatic classification of chickpea varieties using computer vision techniques. Agronomy 9(11):672. https://doi.org/10.3390/agronomy9110672
Tang Y, Cheng Z, Miao A, Zhuang J, Hou C, He Y, Chu X, Luo S (2020) Evaluation of cultivar identification performance using feature expressions and classification algorithms on optical images of sweet corn seeds. Agronomy 10(9):1268. https://doi.org/10.3390/agronomy10091268
Ali A, Qadri S, Mashwani WK, Brahim Belhaouari S, Naeem S, Rafique S, Jamal F, Chesneau C, Anam S (2020) Machine learning approach for the classification of corn seed using hybrid features. Int J Food Propert 23(1):1110–1124. https://doi.org/10.1080/10942912.2020.1778724
Ayele NA, Tamiru HK (2020) Developing classification model for chickpea types using machine learning algorithms. Int J Innov Technol Explor Eng 10(1):5–11. https://doi.org/10.35940/ijitee.A8057.1110120
de Medeiros AD, Capobiango NP, da Silva JM, da Silva LJ, da Silva CB, dos Santos Dias DCF (2020) Interactive machine learning for soybean seed and seedling quality classification. Scient Rep 10(1):1–10. https://doi.org/10.1038/s41598-020-68273-y
Wei Y, Li X, Pan X, Li L (2020) Nondestructive classification of soybean seed varieties by hyperspectral imaging and ensemble machine learning algorithms. Sensors 20(23):6980. https://doi.org/10.3390/s20236980
Koklu M, Ozkan IA (2020) Multiclass classification of dry beans using computer vision and machine learning techniques. Comput Electron Agric 174:105507. https://doi.org/10.1016/j.compag.2020.105507
Salam S (2021) Development and evaluation of chickpea classification system based on visible image processing technology and artificial neural network. Innov Food Technol. https://doi.org/10.22104/JIFT.2021.5173.2063
Sonawane S, Mohanty BK (2021) An improved image processing scheme for automatic detection of harvested soybean seeds. J Food Measur Character. https://doi.org/10.1007/s11694-021-01124-0
Velesaca HO, Mira R, Suárez PL, Larrea CX, Sappa AD (2020) Deep learning based corn kernel classification. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops
Taheri-Garavand A, Nasiri A, Fanourakis D, Fatahi S, Omid M, Nikoloudakis N (2021) Automated in situ seed variety identification via deep learning: a case study in chickpea. Plants 10(7):1406. https://doi.org/10.3390/plants10071406
Zhao G, Quan L, Li H, Feng H, Li S, Zhang S, Liu R (2021) Real-time recognition system of soybean seed full-surface defects based on deep learning. Comput Electron Agric 187:106230. https://doi.org/10.1016/j.compag.2021.106230
McDonald LS, Assadzadeh S, Panozzo JF (2021) Images, features, or feature distributions? A comparison of inputs for training convolutional neural networks to classify lentil and field pea milling fractions. Biosys Eng 208:16–27. https://doi.org/10.1016/j.biosystemseng.2021.05.011
Słowiński G (2021) Dry beans classification using machine learning. Proceedings http://ceur-ws.org ISSN, 1613: p. 0073
Zhang J, Dai L, Cheng F (2021) Corn seed variety classification based on hyperspectral reflectance imaging and deep convolutional neural network. J Food Measur Character 15(1):484–494. https://doi.org/10.1007/s11694-020-00646-3
Taspinar YS, Cinar I, Koklu M (2022) Classification by a stacking model using CNN features for COVID-19 infection diagnosis. J Xray Sci Technol 30:73–88. https://doi.org/10.3233/XST-211031
Jogin M, Mohana MS, Madhulika GD, Divya RK, Meghana S, Apoorva S (2018) Feature Extraction using Convolution Neural Networks (CNN) and Deep Learning. In 2018 3rd IEEE International Conference on Recent Trends in Electronics, Information & Communication Technology (RTEICT). https://doi.org/10.1109/RTEICT42901.2018.9012507
Koklu M, Cinar I, Taspinar YS (2021) Classification of rice varieties with deep learning methods. Comput Electron Agric 187:106285
Dhillon A, Verma GK (2020) Convolutional neural network: a review of models, methodologies and applications to object detection. Progr Artif Intell 9(2):85–112. https://doi.org/10.1007/s13748-019-00203-0
Hemanth DJ, Anitha J, Naaji A, Geman O, Popescu DE, Son LH (2019) A modified deep convolutional neural network for abnormal brain image classification. IEEE Access 7:4275–4283. https://doi.org/10.1109/ACCESS.2018.2885639
Bengio Y, Courville A, Vincent P (2013) Representation learning: a review and new perspectives. IEEE Trans Pattern Anal Mach Intell 35(8):1798–1828. https://doi.org/10.1109/TPAMI.2013.50
Chen Z, Ho P-H (2019) Global-connected network with generalized ReLU activation. Pattern Recogn 96:106961. https://doi.org/10.1016/j.patcog.2019.07.006
Ide H, Kurita T (2017) Improvement of learning for CNN with ReLU activation by sparse regularization. In: 2017 International Joint Conference on Neural Networks (IJCNN). https://doi.org/10.1109/IJCNN.2017.7966185
Glorot X, Bordes A, Bengio Y (2011) Deep sparse rectifier neural networks. in Proceedings of the fourteenth international conference on artificial intelligence and statistics. 2011. JMLR Workshop and Conference Proceedings
Aslan MF, Sabanci K, Durdu A, Unlersen MF (2022) COVID-19 diagnosis using state-of-the-art CNN architecture features and Bayesian Optimization. Comput Biol Med 142:105244
Pan SJ, Yang Q (2009) A survey on transfer learning. IEEE Trans Knowl Data Eng 22(10):1345–1359. https://doi.org/10.1109/TKDE.2009.191
Pan SJ, Yang Q (2010) A survey on transfer learning. IEEE Trans Knowl Data Eng 22(10):1345–1359. https://doi.org/10.1109/TKDE.2009.191
Martinez JB, Gill G (2019) Comparison of pre-trained vs domain-specific convolutional neural networks for classification of interstitial lung disease. In 2019 International Conference on Computational Science and Computational Intelligence (CSCI). https://doi.org/10.1109/CSCI49370.2019.00188.
Dong N, Zhao L, Wu CH, Chang JF (2020) Inception v3 based cervical cell classification combined with artificially extracted features. Appl Soft Comput 93:106311. https://doi.org/10.1016/j.asoc.2020.106311
Szegedy C, Wei L, Yangqing J, Sermanet P, Reed S, Anguelov D, Erhan D, Vanhoucke V, Rabinovich A (2015) Going deeper with convolutions. In 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR). https://doi.org/10.1109/CVPR.2015.7298594
Mednikov Y, Nehemia S, Zheng B, Benzaquen O, Lederman D (2018) Transfer representation learning using Inception-V3 for the detection of masses in mammography. In 2018 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC). IEEE. https://doi.org/10.1109/EMBC.2018.8512750
Simonyan K, Zisserman A (2014) Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556
Banan A, Nasiri A, Taheri-Garavand A (2020) Deep learning-based appearance features extraction for automated carp species identification. Aquacult Eng 89:102053. https://doi.org/10.1016/j.aquaeng.2020.102053
Koklu M, Cinar I, Taspinar YS (2022) CNN-based bi-directional and directional long-short term memory network for determination of face mask. Biomed Signal Process Control 71:103216. https://doi.org/10.1016/j.bspc.2021.103216
Theckedath D, Sedamkar RR (2020) Detecting Affect States Using VGG16, ResNet50 and SE-ResNet50 Networks. SN Comput Sci 1(2):79. https://doi.org/10.1007/s42979-020-0114-9
Singh D, Taspinar YS, Kursun R, Cinar I, Koklu M, Ozkan IA, Lee H-N (2022) Classification and analysis of pistachio species with pre-trained deep learning models. Electronics 11(7):981
Zhang X, Zou J, He K, Sun J (2015) Accelerating very deep convolutional networks for classification and detection. IEEE Trans Pattern Anal Mach Intell 38(10):1943–1955
Carvalho T, De Rezende ER, Alves MT, Balieiro FK, Sovat RB (2017) Exposing computer generated images by eye’s region classification via transfer learning of VGG19 CNN. In 2017 16th IEEE International Conference on Machine Learning and Applications (ICMLA). IEEE. https://doi.org/10.1109/ICMLA.2017.00-47
Maalouf M (2011) Logistic regression in data analysis: an overview. Int J Data Anal Tech Strat 3(3):281–299
Rymarczyk T, Kozłowski E, Kłosowski G, Niderla K (2019) Logistic regression for machine learning in process tomography. Sensors 19(15):3400
Koklu M, Kursun R, Taspinar YS, Cinar I (2021) Classification of date fruits into genetic varieties using image analysis. Math Probl Eng 2021:4793293. https://doi.org/10.1155/2021/4793293
Koklu M, Unlersen MF, Ozkan IA, Aslan MF, Sabanci K (2022) A CNN-SVM study based on selected deep features for grapevine leaves classification. Measurement 188:110425
Scholkopf B, Smola AJ (2018) Learning with kernels: support vector machines, regularization, optimization, and beyond. MIT Press, USA
Cinar I, Koklu M, Tasdemir S (2020) Classification of raisin grains using machine vision and artificial intelligence methods. Gazi Mühendislik Bilimleri Dergisi (GMBD). 6(3):200–209. https://doi.org/10.30855/gmbd.2020.03.03
Ballabio D, Grisoni F, Todeschini R (2018) Multivariate comparison of classification performance measures. Chemom Intell Lab Syst 174:33–44. https://doi.org/10.1016/j.chemolab.2017.12.004
Ropelewska E, Slavova V, Sabanci K, Aslan MF, Cai X, Genova S (2022) Discrimination of onion subjected to drought and normal watering mode based on fluorescence spectroscopic data. Comput Electron Agric 196:106916
Fawcett T (2006) An introduction to ROC analysis. Pattern Recogn Lett 27(8):861–874. https://doi.org/10.1016/j.patrec.2005.10.010
Witten IH, Frank E (2002) Data mining: practical machine learning tools and techniques with Java implementations. ACM SIGMOD Rec 31(1):76–77
Kuncheva LI (2014) Combining pattern classifiers: methods and algorithms. Wiley, USA
Ropelewska E, Sabanci K, Aslan MF (2022) Authentication of tomato (Solanum lycopersicum L.) cultivars using discriminative models based on texture parameters of flesh and skin images. Eur Food Res Technol 248:1959–1976
Berrar D (2018) Cross-validation. Encyclopedia of bioinformatics and computational biology, vol 1. Elsevier, pp 542–545. https://doi.org/10.1016/B978-0-12-809633-8.20349-X
Xu Y, Goodacre R (2018) On splitting training and validation set: a comparative study of cross-validation, bootstrap and systematic sampling for estimating the generalization performance of supervised learning. J Anal Test 2(3):249–262. https://doi.org/10.1007/s41664-018-0068-2
Funding
This project was supported by the Scientific Research Coordinator of Selcuk University with the project number 22111002.
Author information
Authors and Affiliations
Contributions
OZKAN and KOKLU collected data. CINAR conducted literature search. KURSUN conducted material and method research. TASPINAR and DOGAN tested methods for work. CINAR, KURSUN, DOGAN and TASPINAR prepared the article. OZKAN and KOKLU revised and edited the article. All authors agree and approve the final version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Compliance with ethics requirements
This study does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Taspinar, Y.S., Dogan, M., Cinar, I. et al. Computer vision classification of dry beans (Phaseolus vulgaris L.) based on deep transfer learning techniques. Eur Food Res Technol 248, 2707–2725 (2022). https://doi.org/10.1007/s00217-022-04080-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00217-022-04080-1