Abstract
Viruses are changing the biosensing and biomedicine landscape due to their multivalency, orthogonal reactivities, and responsiveness to genetic modifications. As the most extensively studied phage model for constructing a phage display library, M13 phage has received much research attention as building blocks or viral scaffolds for various applications including isolation/separation, sensing/probing, and in vivo imaging. Through genetic engineering and chemical modification, M13 phages can be functionalized into a multifunctional analysis platform with various functional regions conducting their functionality without mutual disturbance. Its unique filamentous morphology and flexibility also promoted the analytical performance in terms of target affinity and signal amplification. In this review, we mainly focused on the application of M13 phage in the analytical field and the benefit it brings. We also introduced several genetic engineering and chemical modification approaches for endowing M13 with various functionalities, and summarized some representative applications using M13 phages to construct isolation sorbents, biosensors, cell imaging probes, and immunoassays. Finally, current issues and challenges remaining in this field were discussed and future perspectives were also proposed.
Graphical Abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In 1985, Professor George Smith first invented phage display technique [1]. By inserting foreign DNA into the genome of phages, endogenous fusion proteins can be displayed on the outer surface of phages. A phage display library can be generated by incorporating a mixture of such phages, each displaying different proteins. Ever since its invention, phage display technique has enabled various in vitro evolution for the selection of protein/peptide with desired properties. This powerful technique won Nobel Prize in 2018, bringing more knowledge about M13 phage to the public [2]. Now, we know that, as the most frequently used phages for constructing phage display library, M13 phage acts more than just basic elements for library construction; it has become a versatile building block for various applications including sensing, imaging, therapeutics, and energy harvesting. As a type of bacteriophage, M13 phage specifically infects bacteria; thus, it is safe for humans, which brings benefit for applying M13 phages in the field of in vivo imaging and therapeutics. In this review, we mainly focused on the application of M13 phages in the field of analytical chemistry. The chemical/genetical manipulations of other virions and their applications in nanoscience, medicine, or material engineering are out of the scope of this review. The readers can find related content in some excellent reviews [3,4,5]. We emphasized the advantages of M13 phage either in its unique structure or its functionalization versatility in improving the analytical performance of the phage-based method. We also addressed the limitations of the currently developed method and proposed possible trends in this field.
Generalities of M13 phage
Structure of M13 phage
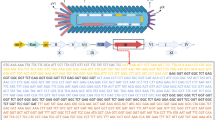
M13 phage is a type of non-lytic filamentous bacteriophage, with a diameter of ~ 6 nm and a length of ~ 880 nm (Fig. 1A). Its well-defined genome was packaged in five capsid proteins (pIII, pVI, pVII, pVIII, pIX) and determines the length of M13 phage [6]. Among the five capsid proteins, minor capsid proteins pIII and pVI are capped on one end, while pVII and pIX are on the other. Each minor capsid protein has 3–5 copies. Major capsid protein pVIII consists of about 2700 copies, which accounts for 98% of the whole mass of M13 phage and contains 50 amino acids (AA). Each pVIII is composed of three domains: positively charged domain (40–50 AA) that interacts electrostatically with phage genomic DNA, intermediate hydrophobic domain (21–39 AA), and the N-terminal domain (1–20 AA). A total of 2700 copies of pVIII protein are helically wrapped around phage DNA through electrostatic interaction between phage genomic DNA and the positively charged domain of pVIII protein, leaving the M13 surface with negative charge.
A Illustration of the structure of M13 phage and the amino acid sequence of pVIII protein. B Schematic showing the infection and production process of natural M13 phage. C Illustration of the mechanism of phage vector-based display system. D Illustration of the mechanism of phagemid-based display system
Minor capsid protein pIII is responsible for the recognition and infection of host cells. Although pIII has only 3 ~ 5 copies, it is one of the most widely used sites for the display of polypeptides as its unconstrained protein structure allowed insertion of exogenous polypeptides of more than 100 AA [7]. Besides capsid proteins, there are several functional proteins responsible for the replication and assembly of M13 phages. As their functions are out of the scope of this review, readers might find more details in some excellent book chapters [8,9,10,11,12,13]
Genetic engineering of M13 phage
One unique advantage of phages over other artificial biomaterials is its ability to display protein/peptide on the desired coat proteins through genetic modification. There are mainly two ways to genetically modify M13 phages, surface display via phage vector or through phagemid.
Phage vector-based display system
Unlike genome DNA of M13 phage, the phage vector is a closed double-stranded circular DNA molecule (Fig. 1C). After molecular biological transformation in vitro, phage vectors can be easily purified and transformed into host cells according to general protocols. A phage vector successfully transformed into a host cell replicates to produce a complete phage particle, which contains only single-stranded DNA ( +). Phage vectors overcome the difficulty that ssDNA is not suitable as a substrate for most commercial restriction endonucleases or DNA ligases, and enables the DNA of M13 phage to be constructed and recombined by conventional molecular biology methods (site-directed mutagenesis, exogenous recombination, Sanger sequencing, etc.). Diverse phage vectors were developed, aiming to insert peptide on pVIII or pIII, in the manner of either mono-display or multiple-display. One of the most frequently used commercially available phage vectors, M13KE, derived from the M13mp vector series [14], was originally designed for displaying peptides on pIII and can also be facilely recombined for inserting peptides in other desired sites. The insertion of gene fragments into M13KE vector for displaying short peptides on pVIII was described in detail as a general protocol by Seung-Wuk Lee’s group [15].
Angela M. Belcher’s group has been engaged in engineered M13 phage for nanomaterials with elevated performance used for energy, the environment, and medicine for several decades. Capsid protein pVIII has been engineered to alter charge, add binding sites for metals or nanomaterials, or enable protein–protein interactions to direct their self-assembly into defined structures. For example, they recombined M13 vector to display tetraglutamic acid (EEEE-) at the N-terminus of the pVIII protein [16]. The resultant phage particles had more negative surface charge, thus interacting more efficiently with positively charged moieties, such as positively charged iron oxide nanoparticles. On this basis, the gene-encoded SPARC-binding peptide was inserted into the coding region of pIII protein of pVIII-EEEE M13 for in vivo targeting of prostate cancer [17].
Phagemid vector-based display system
While using phage vectors to display exogenous peptides offers once-and-for-all advantages, the method suffers from several fetal drawbacks. First, cloning exogenous DNA into the M13 phage vector carries the risk of deletion mutations, and the longer the length of the exogenous DNA fragment, the higher the instability of this recombinant phage vector. Second, engineered phages that inserted with large exogenous gene fragments grow much slower than those inserted with smaller ones or wild-type phages, which is hard to survive under the pressure of natural selection, thus difficult to be propagated in large scale with high purity. These drawbacks can be partially solved by phagemid vector-based display system.
Phagemid is a hybrid vector that carries both the M13 and plasmid origin of replication, and the recombinant coat protein gene. Phagemids can be conveniently propagated as plasmids. Because the phagemid lacks the phage protein genes required to produce a complete phage, cells bearing a phagemid must be infected by a helper phage that encodes proteins required to replicate and package phagemid DNA into an M13 phage particle (Fig. 1B, D). Of even greater interest is that phagemids can incorporate more complex sequences of DNA (as long as 10 kb) or larger fragments of protein than traditional phage vectors [18, 19]. This advantage not only facilitates the convenient fabrication of phage display libraries, especially phage display antibody libraries [20], but also promoted large-scale production of ssDNA [18], for instance, DNA origami. In addition, by altering the size of the phagemid, the length of M13 nanofibers can be finely tuned from 50 to 1300 nm, which was used for constructing stiffness-tunable nanofoams and ultra-small M13 for in vivo targeted glioblastoma imaging and therapy [21, 22].
Although phagemid display system achieved success in constructing phage display libraries, it may not be the perfect choice for fabricating M13-based functional materials due to the low yield of recombinant M13. During phage particle packaging, wild-type pVIII protein from helper phage would compete with pVIII-fusion protein. This leads to ~ 90% of the resultant phage particles to be “bald phages” without fusion protein [23]. Several modified helper phages were developed to solve this issue. One of the most successful examples is a helper phage named hyperphage®. As the pIII gene of the hyperphage® genome is incomplete, only when the plasmid coexist can a complete phage particle be produced [6]. In this way, the fusion protein can be displayed in all five copies of pVIII with high yield. By using the hyperphage® for recombinant M13 rescue, M13 bearing EGFR-targeting peptide (epidermal growth factor receptor targeting peptide) on pIII was fabricated, which were further decorated with photosensitizing molecules for anticancer photodynamic therapy [24].
Chemical functionalization of M13 phage
From a chemist’s point of view, various types of amino acids contained in the proteinaceous surface of M13 phage provide various reactive sites for bio-orthogonal chemical reactions [25]. Typically, several amino acids are employed as target residues such as N-terminal alanine, lysine, aspartic/glutamic acid, cysteine, N-terminal serine/threonine, and tyrosine. By reacting with these reactive sites, M13 phages can be functionalized with DNA strands, antibodies, nanoparticles, fluorophores, and drugs for wide applications including biosensing, biomedicine, issue generation, and even energy harvesting (Fig. 2).
Natural amino acid reaction
Among them, -NH2 groups in N-terminal alanine and lysine (Lys8) on pVIII protein are the most frequently used reactive sites for M13 functionalization, and NHS chemistry is extensively used as it is efficient and does not require harsh reaction conditions. NHS ester is formed by carbodiimide activation of carboxylate molecules. NHS ester activated cross-linkers and labeled compounds react with primary amines to form stable amide bonds under physiological to weakly alkaline conditions (pH 7.2 to 9). For example, Xiong’s group used a NHS ester named N-succinimidyl 3-(2-pyridyldithio) propionate (SPDP) to react with the amine groups on M13 surface and obtained thiol modified M13 phage. The thiolated M13 was further coupled with AuNPs@Ag to endow M13 with catalytic ability, which significantly improved the sensitivity of the immune sensor [26]. Similarly, Dirk Rothenstein’s group used a heterobifunctional NHS ester cross-linking agent (sulfosuccinimidyl (4-iodoacetyl) aminobenzoate (sulfo-SIAB)) as an intermediate to covalently conjugate urease to phage and obtained a reliable and easy-to-use tool for enzymatic catalytic application [27]. Except for the widely used NHS chemistry, amine group could also be functionalized through isothiocyanate or succinimidyl formylbenzoate.
In general, the reactive carboxyl groups on the pVIII protein were provided by glutamate and aspartate side chain (Glu2, Glu20, Asp4, Asp5). Those carboxyl groups usually react with activated carbodiimide cross-linker (e.g., 1-ethyl-3-(3- (dimethylamino) propyl) carbodiimide hydrochloride (EDC) or dicyclohexylcarbodiimide (DCC)) to produce O-acylisourea intermediate, followed by reacting with amino groups to form stable amido bond under the mild acid environment. For example, Itai Benhar and co-workers developed a kind of nanodrug of phage-dox via the coupling of the carboxyl side chain of M13 phage with the primary amine of neomycin to remarkably enhance the drug-carrying ability and improve the targeting performance of the nanoparticle towards cells [28,29,30]. The pVIII protein of IgG-targeting M13 phage was also chemically modified with thiol group for further AuNP conjugation using EDC and cysteamine for amplified IgG detection via visible plasmon shifts of AuNPs [31].
The phenol groups on two tyrosine residues (Tyr 21 and Tyr 24) of the pVIII protein are reactive with azo salts. The use of chemoselective modification of tyrosine residues requires incorporation within the N-terminal hydrophilic domain for chemical access. For example, Mao’s group prepared photo-responsive organic nanowires Azo-M13 by using M13 phage as a template to genetically display tyrosine-end peptide on the surface of pVIII protein and specifically react with aromatic amines-functionalized dyes via diazotization reaction. The Azo-M13 complex showed excellent reversible photo-response performance [32]. Wang’s group modified M13 phages with alkynylated group via the reaction between phenol groups on the pVIII protein and alkynylated diazo. In the presence of Cu2+, the alkynylated M13 was further conjugated with folic acid via click chemistry for cancer cell imaging [33].
Cysteine is often used as alkylation site for electrophilic halides or maleimides. However, pVIII protein does not contain cysteine. Cysteine only presents in the minor capsid proteins pIII, pVII, and pIX to form disulfide bonds for maintaining protein structures. Through genetic engineering, cysteine can be embedded in the capsid proteins to produce reaction handles [34].
N-terminal serine and threonine, which are absent in wild-type M13 phage, also need to be genetically displayed for oxidation into aldehyde by NaIO4. As 20–30% of the peptide-displayed M13 phages in the commercially available random peptide libraries contain N-terminal serine and threonine, glycopeptide libraries were prepared by NaIO4 oxidation of the N-terminal serine and threonine, which further undergone oxime ligation to introduce glycan [35].
Non-canonical amino acids (ncAAs) integration
As the same kind of reactive group may exist in more than one type of amino acid residues on M13 capsid protein, cross-reactions would inevitably occur, which would greatly reduce the modification efficiency of the desired sites. To realize site-specific modification, non-canonical amino acids (ncAAs) can be integrated into the capsid protein of M13 phage [36, 37]. Selenocysteine (Sec), the 21st amino acid, was displayed on the pIII protein of M13 phage using a natural selenocysteine opal suppressing tRNA. As Sec displays stronger nucleophilicity and reactivity than Cys at physiological pH, it allowed selective tethering of small molecules to Sec in a seleno-pIII displaying phage [38, 39]. Later, Schultz group reported a series ncAA systems incorporating phenylalanine-derived ncAAs into M13 phages to label proteins expressed on phage through azide-alkyne cycloadditions and Staudinger ligations [40, 41]; they also used the similar system to evolve single-chain antibody variable fragments (ScFvs) with chemical warheads and proteins that chelate metal ions [42,43,44]. While this ncAA displaying system only incorporates a single type of ncAA in response to a single amber codon in the gene of interest, Chin’s group developed a system enabled encoding of multiple, structurally and functionally diverse ncAAs [45].
Recently, a Nϵ-acryloyl-lysine (AcrK) was genetically encoded in a phage-displayed peptide library for cyclization with a pre-installed cysteine via a proximity-driven Michael addition reaction between cysteine and AcrK, solving the problem encountered in cysteine conjugation [46].
Phage display
An important way to get M13 phages displaying exogenous protein/peptide is through phage display technique, or so-called biopanning [47]. Phage display technique, the Nobel Prize-winning technique first invented in 1985, is an evolutional approach via the high-throughput selection of peptide ligands of diversity, which has become an indispensable tool for exploring specific biochemical molecular interactions [48]. To select specific ligands through phage display, a broad and efficient method of affinity enrichment, commonly referred to as biopanning, is widely used, including multiple rounds of positive selection, counter selection, washing, elution, and amplification. With the increase of biological screening cycles, high-affinity ligands are gradually enriched. The sequence of these high-affinity ligands can be obtained by DNA sequencing of the corresponding phage monoclone [49]. For instance, an angiogenin-targeting phage was selected in vitro from a random 12-mer peptide library for tumor angiogenesis suppressing [50]. In vivo biopanning through the intraperitoneal injection of the peptide library facilitated the discovery of peptides able to target drug uptake by a vast array of neurons of the autonomic nervous system (ANS) [51].
While conventional in-plate biopanning suffers from drawbacks of high repetition, time-consuming process, labor-intensive operation, and risks of contamination, microfluidic technology offers several unique advantages to solve these problems. Zhu’s group has developed several microfluidics-based biopanning platform for rapid and efficient selection of target-binding phages. By using a droplet microfluidic system, single M13 phage was embedded into one droplet for phage amplification, target-beads interactions, and high-affinity phage identification, which can obtain target-binding phages within 24 h (Fig. 3A) [52]. They further developed a molecular selection system “SCOPE” using a deterministic lateral-displacement (DLD) microfluidics, aiming at achieving highly efficient and affinity-discriminated molecular selection. The collision of phage-bound target beads effectively removes phages with low affinity. By combining the collision-based force effect with the two-dimensional separation-based concentration effect to increase the biopanning stringency, phages with high-affinity can be screened within only 6 h (Fig. 3B) [53]. Professor Shanna O’ Kelley’s group has developed a biopanning platform “Cellect” based on their previously fabricated immunomagnetic cell sorting microfluidic chip, aiming at identifying high-affinity binders against challenging targets using minimal rounds of selection. As shown in Fig. 3C, cells presenting target antigens were mixed with background cells to mimic the in vivo binding environment. The biopanning stringency can be tuned by changing the ratio of cell types. With the assistance of machine learning and next-generation sequencing (NGS), binders with picomolar affinity can be identified with only two rounds of biopanning [54].
Microfluidic-based biopanning strategies for the screening of target M13 bacteriophages. A Mechanism of droplet microfluidic-based double monoclonal display (dm-display) system. Copyright 2021 Royal Society of Chemistry [52]. B Scheme of the SCOPE system-based phage screening for micropillar array and particle tracing. Copyright 2022 National Academy of Science [53]. C Schematic illustration of the μCellect methodology. Copyright 2022 American Chemical Society [54]
Engineered phages for elevated analytical performance
Phages are such a unique nanomaterial that functional groups can either grow (genetic engineering) or decorated (chemical modification) on their surfaces. While genetic engineering can achieve site-specific functionalization of particular types of coat protein, chemical modification possesses the advantage to further expand the functionality of phages beyond natural peptide and proteins. By taking merit of the genetic engineering and chemical modification, different types of coat protein can be manipulated to possess desired functionality, endowing M13 phages as a multifunctional sensing platform with various function regions conducting their functionality without mutual disturbance. Due to the unique feature of M13 phages, such as filamentous morphology and flexibility, particular benefit has been gained engaging engineered M13 phages in analytical applications. In the following, some representative applications to use M13 phages in the construction of analyte isolation sorbent and the fabrication biosensor, as well as in immunoassay and cell imaging will be introduced, with working principles, advantages/disadvantages and benefits from M13 phages discussed in detail.
Analyte isolation and separation
With the unique feature of the filamentous morphology and the orthogonal reactivity, M13 phages can be used as an ideal alternative for the construction of immunosorbents for analyte isolation. The target binding ability can be obtained by decorating the binding ligands (antibodies, aptamers, etc.) on the surface of phages through chemical conjugation. In order to achieve better analyte separation, phages were usually immobilized on a solid substrate. Gil U. Lee et al. [55] have systematically evaluated several coupling strategies to immobilize M13 phage on the surface of magnetic beads and found that chemical coupling through EDC/NHS reaction or neutravidin-biotin conjugation results in relatively larger phage coverage, but in a side-on manner. On the other hand, by genetically modifying M13 phage to display 6His on the pIII protein, the end-on anchoring of M13 can be realized through 6His-NTA interaction with the advantage of strong yet reversible assembly. The end-on manner also facilitated the construction of bioinspired architectures mimicking the cellular threadlike structures and allows high degree of conjugation with functional molecules. By displaying a 14-mer biotin acceptor peptide on the pIII protein, M13 phages were biotinylated with the assistance of biotin ligase BirA for end-on anchor of M13 on streptavidin-functionalized microbeads. The pVIII protein was further functionalized with antibodies through Staudinger ligation for constructing immunoassays. The resultant phage-decorated microbeads outperformed the non-phage counterparts in terms of both sensitivity and selectivity. The low non-specific binding of the phage-decorated microbeads was mainly due to the gold layer coating, whereas the improved sensitivity was attributed to the increased antibody loading and enhanced interaction between target molecules and flexible phages [56].
Similarly, anti-HER2 antibody was tethered to every amine group the pVIII protein using 6-arm PEG15000-NHS as linkers [57]. The antibody-loaded phages were subsequently anchored to the magnetic beads via specific recognition between pIII protein and anti-gp3 antibody modified on the surface of the magnetic beads. Approximately 162,000 antibody molecules were loaded on M13 anchored magnetic beads, which is 2.4-fold higher than that of conventional immune-magnetic beads. These nanotentacle-structured magnetic beads had high affinity towards HER-2 positive cancer cells, with high capture purity and efficiency. The author claimed that the increased target binding affinity was mainly due to the multivalent interaction between the antibody molecules on the flexible M13 and receptors on the target cell. According to the estimation provided by the author, ~ 157 antibody molecules were functionalized on M13 phages. Considering the distance between two N-termini of the pVIII protein is 2.4–3.2 nm and 2700 copies of pVIII proteins on each M13, the distance between two antibody molecules was ~ 40–54 nm. For such a long distance, multivalent effect might not be predominant enough for multiple antibody molecules to bind with the same receptor molecule/cluster on cell surface. Aiming at taking advantage of the flexibility feature of M13 to achieve enhanced multivalent interaction, we recently constructed a dual-functionalized M13 phages, with pIII protein genetically displaying 6His tag for end-on anchoring onto magnetic beads, and pVIII protein conjugated with MUC1-tageting aptamers through click reaction. As the size of aptamers was only 1/10 of that of antibodies, more than 600 aptamer molecules can be conjugated on the surface of M13. The aptamers target the variable number tandem repeat (VNTR) region of MUC1, where the 200–500-nm-long MUC1 possesses about 20–120 VNTR repeats. This indicated that the density of aptamer on M13 surface fitted well with that of VNTR on MUC1 molecule. With nanoscale matching between VNTR repeats and corresponding aptamers, sub-micron-scale matching between MUC1 and M13 nanofibers, and the topological interaction between cancer cells and dual-functionalized M13-anchored magnetic beads, our cancer cell isolation platform resulted in six magnitude-improvement in cancer cell binding affinity with respect to that of free aptamer.
While the decoration of binding ligands requires complicated reaction procedure, biopanning from the peptide library provides easier way to obtain target binding ability. Our group has developed a simple and effective Cr(III) preconcentration approach using Cr(III)-binding-M13 as recognition element. The Cr(III)-binding-M13 were obtained through repeated biopanning from a 7-mer random peptide library. Besides conventional positive biopanning, negative biopanning against blank resin and foreign metal ions were added to improve the binding selectivity (Fig. 4A). For the ease of sample separation, the Cr(III)-binding-M13 were further immobilized on microbeads through electrostatic interaction. By eluting the retained Cr(III) with HNO3, and measuring the eluted Cr(III) by ICP-MS, a limit of detection (LOD) of 15 ng/L was achieved for Cr(III) [58].
Representative examples of M13-based recognition strategies. A Graphic of the biopanning procedure to obtain the Cr(III) binding phages. Copyright 2015 American Chemical Society [58]. B Chimeric M13 phage decorated with AuNPs for detection of bacteria. Copyright 2019 American Chemical Society [71]. C Construction of TC-phage-FlAsH structure for bacterial detection. TC-tag displaying M13 phage recognized and infected the host bacteria and then labeled with the fluorescent dye FlAsH-EDT2. Copyright 2014 Wiley–VCH GmbH [72]. D Schematic illustration of PhaNGS approach. The phage particles were designed to specifically binding to target proteins and identified by unique sequence through NGS. Copyright 2018 National Academy of Science [78]
The analysis of low abundance proteins in serum samples faces great challenge due to the enormous dynamic range of protein concentrations. Yu-Kui Zhang’s group developed a facile protein abundance equalization approach using scFv displaying M13 phage library (scFv@M13) [59]. As an scFv displaying library is an antibody pool with a diversity of up to 106, it is efficient for the binding of all proteins in complex samples. By immobilizing scFv@M13 onto magnetic microspheres through glutaraldehyde cross-linking, the scFv@M13 immobilized magnetic microspheres (scFv@M13@MM) were firstly co-incubated with human serum to capture the target proteins, and then were separated and enriched by protease release and digestion for SDS-PAGE and MS analysis. Excessive high abundant proteins cannot be retained on scFv@M13@MM, whereas low abundant proteins were gradually enriched by overloading. The separation efficiency of proteins in serum samples by scFv@M13@MM reached 100%, which could effectively remove high abundance proteins, reduce protein concentration differences in serum, and identify more kinds of low abundance proteins. Compared with peptide library, scfv@M13 has stronger strong binding capacity and high specificity and can be regenerated to provide sufficient binding sites.
Immunoassay
Immunoassay is the most predominant form of phage-based analysis approaches, where the target-binding M13 acts either as “capture antibody” or as “signaling antibody.” The M13-based capture antibody leverages the multivalency of the pVIII display system for enhanced analyte capture, whereas the M13-based signaling antibody benefits from the 5:2700 molecular ratio between pIII and pVIII to achieve signal amplification.
M13 phage as capture antibody
In immunoassays where M13 phages act as capture antibodies, sandwich type assay was the most frequently adopted, in which M13 phages that can specifically bind to the target (usually immobilized on a solid surface) first capture the target molecules and the latter were then labeled by suitable probes for signal output. Biopanning from peptide libraries is the most straightforward way to obtain target-binding M13 phages. By combining the phage peptide library screening technology with enzyme-linked immunosorbent assay (ELISA), the phage ELISA method was developed. Liu’s group [60] screened the f8/8 phage peptide library to obtain a phage monoclone with pVIII protein displaying ERNSVSPS with high specificity towards free-PSA, and then introduced anti-PSA-mAb to recognize the free-PSA captured by the phage. Benefiting from multivalency of pVIII-displaying phage, a LOD of 0.16 ng/mL was achieved together with a detection range between 0.825 and 165 ng/mL. Based on the conventional pIII-based display system phagemid pComb3XSS, Hua et al. [61] constructed cyclic 8-, 9-, and 10-residue peptide libraries using phagemid pCom-pVIII, which can not only achieve high-density display of peptides but also avoid potential loss of diversity of peptide libraries caused by restriction endonucleases. Through blended biopanning of cyclic peptide libraries, phages with high specificity towards clothianidin were obtained, which were successfully used to develop competitive phage ELISA and non-competitive phage ELISA. Besides conventional enzyme-based colorimetric signal output, the M13 phage with specific recognition ability could be conjugated with nanoparticle to build SERS sensing platform. A phage containing a highly specific peptide sequence (RKIVHAQTP) for U937 cells was obtained by biopanning from a 9-mer pVIII M13 random peptide library. Phage-AgNPs network was subsequently formed for the label-free Raman analysis of U937 cell [62]. Similarly, by biopanning from a 9-mer peptide library, three types of phages targeting three different pathogens were obtained, which were further immobilized on magnetic beads for analysis with Raman spectrometry [63].
Besides biopanning, target-binding M13 phages can also be constructed by phage display. For instance, M13 phage was engineered to display paraquat-binding peptide (BPWHW) on pVIII proteins for SERS detection of pesticide residues of paraquat [64]. By simply mixing the engineered M13 phage with silver nanowires (AgNWs) and vacuum filtered onto glass fiber paper (GFFP), a paper-based SERS sensing material was constructed. As AgNWs were stacked on top of GFFP to form multiple hot spots, the interaction between phages and the target pesticide residues can be reflected by SERS signals. This paper-based material is easy to prepare and can be used for in situ detection. In another example, a streptavidin-binding peptide was displayed on the pVIII protein of M13 phages for the label-fee optical detection of streptavidin [65]. Polymer biosensor PVA/M13 fibers were prepared in a one-step process using a blend near-field electrospinning technique (blend NFES), with the streptavidin-specific M13 phage as affinity capture agent, and rhodamine 6G as a high-yield fluorescent signal source. The resonance cavity was excited with a 532-nm continuous laser focused through the microscope and the spectral response was observed, causing a change in the whispering gallery mode resonance peak (i.e., red-shift) when streptavidin was present, thus enabling real-time monitoring of streptavidin levels.
M13 phage as signaling antibody
By using M13 as signaling antibody, the pIII protein was usually displayed with target-binding peptide, whereas the pVIII protein was functionalized with signal reporting molecules like fluorescence tag, metal nanoparticles and DNA sequences. While each M13 can bind few target molecules through 3–5 copies of pIII protein, hundreds to thousands of signal reporting molecules on the phage surface can be labeled, which greatly amplified the signal. Jennifer N. Cha’s group has reported a series of M13-based signal amplification system. They first functionalized the pVIII protein of an IgG-targeting M13 phage with thiol groups using EDC and cysteamine [31]. Due to the high avidity between thiol and gold, the addition of AuNPs into thiolated M13 results in its aggregation which can be used as a sensitive signal output. However, the reactivity of -COOH on pVIII is relatively lower than -NH2; this chemical modification resulted in only 136 thiol groups functionalized per phage. After that, they modified the pVIII protein with thiol groups by NHS ester reaction with -NH2 using SPDP. This led to a conjugation of 259 ± 19 maleimide functionalized DNA onto each M13. The DNA-phage was used to hybridize with AuNPs-DNA for quantification of targeted proteins based on colorimetric and nanoarray analysis [66]. Although EDC/NHS coupling method is the most frequently used bioconjugation method, it may cause intra-virus or inter-virus cross-linking that would reduce the DNA linking efficiency. Aiming at solving this issue, they incorporated benzaldehyde into the pVIII protein of M13 by NHS ester reaction, and then coupled with hydrazide derived DNA via the hydrazone bond. With this method, each phage can be conjugated with 460 ~ 675 DNA molecules, significantly improving the fluorescence signal [67]. In order to further enhance the signal, the DNA-phage conjugation system of hydrazone bonds was further labeled with DNA-functionalized Au@Ag SERS tag through DNA hybridization to detect target proteins. The Raman intensity of the DNA-phage system was nearly 75 times higher than that of the DNA-antibody, which greatly improved the sensitivity [68]. Our group has obtained a S. aureus-binding phage pSA-1 from the 12-mer peptide library and decorated pSA-1 with AuNPs in situ. The labeling of Raman active molecule DTNB endowed AuNPs with SERS activity, facilitating rapid and sensitive detection and simultaneous inactivation of S. aureus [69].
Besides colorimetric and SERS signal output, chemiluminescence (CL) was also incorporated in phage ELISA with advantages of high sensitivity. Very recently, a SARS-CoV-2-specific phage displaying the peptide sequence NFWISPKLAFAL was biopanned from a pIII phage library using the S protein of SARS-CoV-2 as a target protein [70]. While the pIII protein was used for target capture, the pVIII protein provided multiple binding sites for high-density modification of the signaling antibody anti-M13-HRP mAb. Hence, a sandwiched phage-based enzyme-linked chemiluminescence immunoassay (ELCLIA) assay was established, which can detect SARS-CoV-2 down to 78 pg/mL and can be used to detect heat-inactivated SARS-CoV-2 pseudovirus (D614G) at 60 TU/mL in 50% saliva, demonstrating the advantages of phage sensing tools that are resistant to high temperatures and can be sensitively detected in complex matrices.
As the naturally existing phage is the result of long-term evolution by nature, the affinity between naturally existing receptor binding proteins (RBP) of phages and the host cells is very tight and specific. Therefore, other than obtaining target-binding phages through biopanning from peptide libraries or genetically displaying target-binding peptide/antibody/nanobody on phages, Irene A. Chen’s group displayed the RBPs for constructing phages with high affinity towards six types of host bacteria (Fig. 4B). These chimeric phages were further thiolated for the binding of AuNPs as signal output, which resulted in sensitive detection limit of down to ~ 100 cells [71].
In addition to the chemical conjugation of signaling moieties, genetic engineering approach can also be used to enable signal labeling. For example, Wu et al. inserted an optimized tetracysteine sequence (TC-tag) into the minor coat protein coding region of M13KE. The displayed TC-tag was displayed on M13 and entered the host cell via infection. Strong fluorescent signal generated upon the binding between TC-tag and fluorescein arsenical helix binder (FlAsH), a membrane-permeant biarsenical dye, facilitating host cell monitoring and viability identification under the detection of flow cytometry (Fig. 4C) [72, 73].
One of the unique features of phages compared with other biomaterials is the correspondence of the genotype and the phenotype. As the peptide is genetically coded and displayed on the phage particle that packed the peptide gene inside, the peptide coding region can be used as signal output for the recognition and quantification of this peptide-bearing phage. By using phage as the antibody-DNA analogue, several phage display mediated immune-PCR (PD-IPCR) methods were developed [74, 75]. For instance, an anti-idiotypic nanobody-phage was used for the construction of a competitive ELISA assay for aflatoxin. Phage DNA was released by heat lysing and the nanobody encoding sequence was used as template for real-time quantitative PCR (qPCR) [75]. A LOD of 0.02 ng/mL was achieved, which is fourfold more sensitive than traditional phage ELISA. In addition to qPCR, several nucleic acid amplification strategies were also successfully integrated into PD-IPCR, including rolling circle amplification (RCA) [76] and loop-mediated isothermal amplification (LAMP) [77]. As the analogue of antibody-DNA, recombinant phages are easier to get, i.e., from phage display peptide library. By combining the NGS technique with phage display, James A. Wells’ Group [78] proposed genetically barcode antibody for phage antibody next-generation sequencing (PhaNGS) for proteomics research. As can be seen from Fig. 4D, antibodies with specific fragments bound to specific targets were first screened from a large library of synthetic Fabs (about 1010 unique sequences). The selected Fab was encoded and displayed on phage particles to construct the Fabs-Phage system. Not only can a single Fab-phage bind to its corresponding antigen-binding site, but the H3 loop of the complementarity determining part (CDR) can also be used as a recognition barcode for special labeling and quantification to achieve multiple quantification of the proteome surface. In this work, 144 preselected antibodies against 44 receptor targets displayed on filamentous phage (Fab-phage) were used to evaluate the changes of proteins and oncogenes. The results were similar to those obtained by flow cytometry and mass spectrometry, indicating the reliability of the study. At the same time, the low cost and hundreds of antibody tags make Fab-phage widely used in high-throughput sequencing. By further integrating this phage barcode with microfluidic system, Shana O. Kelley’s group [79] achieved protein expression analysis on the surface of single captured circulating tumor cell (CTC). In this work, cobalt-based magnetic alloy guides were combined with chips to achieve efficient capture and separation (~ 90%) of CTCs by antibody-functionalized magnetic nanoparticles under magnetic guidance. Fab-phage barcode tags were injected into the chip to monitor their expression in situ in single captured H460 cell. For quantification, unbound Fab-phages were washed by PBS, flushed out of the capture zone and the phage was incubated for a night to propagate. Four single H460 cells individually captured by phage binding onto the chip were sequenced as biological replicates to assess changes in single-cell bound FZD2 levels, indicating a correlation between FZD2 expression and the cellular microenvironment. Aviv Regev’s group [20] also developed a method based on phage high-throughput sequencing nanoantibodies-labeled single-cell mapping sequencing. Phages genetically displayed with nanoantibodies were used for the labeling of various proteins on cell surface and quantified by NGS via the unique hypervariable complementarity determination region 3 (CDR3) barcode. Multimodal single-cell analysis was achieved by integrating the phage-based multiplex protein measurements and chromatin accessibility profiling into the droplet-based single-cell sequencing system.
Sensing
While immunoassay provides general method for analyte detection, it always suffers from complicated operational procedure and time-consuming drawbacks. On the contrary, by measuring the analyte-induced signal transduction (optical or electrochemical signal), M13 phage-based sensing method was regarded to be more attractive due to its advantage of simplicity and rapidness. Nevertheless, due to the coexistence of interferences, sensing method might not be suitable for the direct analysis of samples with complex matrix. As M13 phages can be multi-functionalized, sensors built on M13 phages can be more integrated and simplified.
Optical sensing
Colorimetric sensing
Colorimetric sensing attracted much research attention due to its simplicity and the potential to be developed into naked eye detection method. The well-defined structure and reproducible feature of phages make them ideal building block for the assembly of regular structures. Seung-Wuk Lee’s group reported the self-assembly of genetically engineered M13 phage into target-specific, colorimetric biosensors. The sensors were composed of quasi-ordered phage-bundles with viewing-angle independent color attributed to coherent scattering from the fiber bundle structures. The existence of volatile organic chemicals led to the swelling of structures along with distinct color changes (Fig. 5A). By displaying trinitrotoluene (TNT)-binding peptide motifs on the phages, selective detection of TNT with LOD of down to 300 ppb was achieved [80]. They further assembled M13 phages into liquid-crystalline bundled nanofibers for the rapid and sensitive colorimetric detection of humidity [81]. By further engineering pVIII protein with target responsive short peptides, the discrimination and identification of endocrine disrupting chemicals, cells, and antibiotics were achieved with the assistance of sensing array technique by Jin-Woo Oh’s group [82,83,84].
Optical sensors for specifical determination of biomarkers based on M13 phage particles. A Construction of volatile chemical biosensor inspired by the color-shift characteristic of turkey skin. TNT specific engineered M13 phage could self-assemble into the structure of mimic collagen fibers, which produces different colors through swelling or deswelling to indicate the concentration of TNT in the air. Copyright 2014 Springer Nature [80]. B Construction of M-phage-DNAzymes for miRNA quantification. The HRP-mimic hemin/G-quadruplex DNAzyme could catalyze the oxidation of ABTS2− under the C HCR reaction triggered by target miRNA. Copyright 2022 Wiley–VCH GmbH [86]
While sophisticated skills are required for the assembly of M13 into regular structured alignments, our group reported a simple colorimetric method for Hg sensing by the formation of a phage-nanoparticle network. The M13 phage that specifically binds to Hg2+ was obtained by biopanning from a peptide library, and then, AuNPs were grown in situ on M13 surface to form a phage-AuNPs network. Upon the addition of Hg2+, the reductive sites on M13 surface reduced Hg2+ into Hg0, which further deposited onto AuNPs and induced a color change from light purple to dark pink [85]. Due to the inherent avidity between gold and mercury and the specificity of the Hg2+-binding phage, this approach allows for selective sensing of Hg2+ with high interfere ion tolerance.
Albeit extensively adopted by analytes, the sensitivity of colorimetric sensing is usually limited. By combing enzymatic reactions with colorimetric sensing, the sensitivity can be largely improved. Mao’s group [86] recently constructed a hybrid sensing platform by appointing M13 phage as a bio-carrier, which showed high sensitivity and specificity for circulating microRNA (miRNA) quantification. Artificial enzymes (Hemin/G-quadruplex DNAzymes) were integrated through the HCR reaction triggered by target miRNA (Fig. 5B and C), which subsequently catalyzed the oxidation of ABTS2− and led to an absorbance signal that indicated the level of miRNA. Furthermore, magnetic nanoparticles (MNPs) coated on the side wall of the M13 phage could achieve the separation of miRNA and minimize the background signal in the clinical sample.
Surface plasmon resonance (SPR) sensing
Surface plasmon resonance (SPR) biosensors measure the refractive index changes caused by the interaction between analyte and biosensor surfaces, which has the advantage of rapidness and label-free detection [87]. The surface-displayed peptides of M13 phage can specifically recognize and interact with targets and thus can be used to replace antibodies in SPR sensing applications. For instance, Nitsara Karoonuthaisiri established an SPR assay based on 12-mer peptide-displaying phage to detect the foodborne bacterium Salmonella [88]. Seung-Wuk Lee’s group reported a phage chip for the SPR monitoring of cell proliferation and morphology. Phages were first engineered to display integrin-binding peptide (RGD) or the biotin-like peptide (HPQ) for conjugating growth factors. The recombinant M13 were then self-assembled into monolayer on gold-coated glass substrate. As the interaction between phages and cells affected the SPR optical characteristics, cell responses to physical and chemical cues can be detected [89]. Compared to randomly alignment of the recognition molecules, grating patterns on the surface of the SPR sensor can enhance the efficiency of the SPR sensor. In this regard, Jin-Woo Oh’s group assembled HPQ-displaying M13 into a nematically oriented fibrous structure for the SPR sensing of streptavidin [90]. The regular alignment of M13 fibers effectively enhanced the sensitivity of SPR, with the nematically aligned HPQ phage film perpendicular to the incident light possessing the highest sensitivity. As a powerful technique capable of monitoring the specificity, affinity, and kinetic behavior of interactions between SPR sensor surface and the analyte, the potential of the M13 phage-based SPR sensor to study the interaction between receptor-ligand, protein-DNA, virus-host, and etc. should be further explored, rather than just measure the concentration of the analyte.
Fluorescence sensing
Fluorescent signals are common output signals in sensors due to its high sensitivity. Sung jee Kim’s group developed a fluorescence “turn on” TNT sensor based on the binding strength difference of the TNT-binding M13 phages towards TNT and the quencher. A quantum dot (QD)-recombinant M13 phage hybrid composite film was constructed through layer-by-layer assembly. The presence of the quencher “turned off” the fluorescence of QD, while TNT molecules can displace the quencher on M13 surface and “turned on” the fluorescence. Sensitive detection of TNT can therefore be achieved down to the sub ppb level [91]. Förster resonance energy transfer (FRET) is one of the most extensive sensing mechanisms among fluorescent probes. The appropriate distance between two adjacent pVIII proteins makes M13 phage the perfect platform for conducting FRET sensing. Qian Wang’s group used M13 phage as a scaffold to construct a ratiometric fluorescent nanoprobe based on FRET. Fluorescein isothiocyanate (FITC) and rhodamine B (RhB) were tethered on the N-terminus of M13 phage pVIII protein as FRET donor and acceptor, respectively. -Cyclodextrin (-CD) was first coupled to the N-terminus on M13 surface through carbodiimide chemistry for further anchoring adamantly (Ada)-derived FITC and RhB via the host–guest interaction. As the distance between two neighboring N-termini (2.4 ~ 3.2 nm) is within the Forster distance, FRET process occurred between neighboring Ada-FITC and Ads-RhB. This M13-based FRET platform was further used for the ratiometric pH sensing [92]. Jin-Woo Oh also developed an M13 phage-based on/off FRET sensor for TNT sensing. Water-soluble CdSSe/ZnS nanocrystal quantum dots as the donor and fluorescein isothiocyanate as the acceptor were modified on the surface of M13 phage, respectively. In the presence of TNT, the M13 phage (pVIII displaying TNT-binding peptides) specifically captured TNT, causing a mismatch in the distance between the donor and the acceptor molecules, thereby inhibiting energy transfer and triggering the on/off of FRET [93]. Based on the flexible feature and chemical modifiability of M13 phage, our group recently constructed a dynamic deformable nanointerface for the sensitive detection of pathogen. M13 phages were genetically engineered to display 6His tag for anchoring on the nanointerface. The pVIII proteins were conjugated with multivalent aptamers for the selective pathogen capture. The binding between aptamers and target pathogens released pre-hybrized complementary strands and initiated the RCA reaction for fluorescence signal output. The sway motion of M13 nanofibers effectively accelerated the diffusion of the solution and promoted RCA reaction efficiency, thereby enhancing the sensitivity of this method (Fig. 6) [94].
Dual functional M13 phage for pathogen capturing and sensing. A Fabrication of dynamic deformable nanointerface using flexible M13 phage and biotinylated-aptamer. B Procedure of bacterial capturing and sensing. C Schematic illustration of the significantly enhanced binding affinity and accelerated mass transport through sway motion of flexible M13 phage. Copyright 2022 Wiley-VCH GmbH [94]
Electrochemical sensing
Electrochemical impedance spectroscopy (EIS) is an effective tool for rapid and sensitive monitoring of small changes at the electrode interface. Early detection of discriminating fecal coliforms E. coli XL1-Blue and K12 was realized using a M13 phage-based EIS sensor [95]. M13 phage was functionalized with 3-mercaptopropionic acid to immobilize on AuNPs-posited electrode. The sensor was stable over a wide pH range (3.0–10.0) under high temperature conditions (45 °C) due to its non-cleavable nature. Liu Aihua’s group [96] screened two phages bearing specific octapeptide against free/total prostate-specific antigen (f-PSA/t-PSA) and immobilized them on the surface of gold electrodes, and constructed a pair of label-free EIS biosensors for f-PSA and t-PSA. The phage-based biosensors can be regenerated by elution with Gly-HCl (pH 2.2) and reused for 6 cycles. Similarly, the assembly of ovomucoid-binding phages on a gold electrode resulted in sensitive detection of ovomucoid with a LOD of 0.12 μg/mL [97]. In a recent work, a competitive immuno-electrochemical sensor was fabricated using HRP-labeled phage as competitor for the determination of organophosphorus pesticides (OP). The GCE was modified with doped carbon quantum dots and graphene oxide and then assembled with anti-OP antibody. The phage that mimic OP was decorated with anti-M13 mAb-HRP for signal output. By taking advantage of the unique structure of M13, plenty of anti-M13 mAb-HRP can be decorated, resulting in amplified signal and a LOD of down to 0.003–0.014 ng/mL [98]. Although M13 phage-based electrochemical sensor provided simple and label-free detection for sensitive analysis, two issues should be taken into consideration. First, current studies immobilized M13 phages on the electrode surface in a non-site-specific way, which might lower the effectiveness of M13 phages to bind and interact with the analytes. Second, the immobilization of M13 phages might increase the roughness of electrode surface thus the biofouling issue should be appropriately solved, for instance, by displaying anti-fouling peptide on M13 phages.
Cell imaging
Cell imaging is a characterization method to detect the presence of specific biomarkers in cells and to map their spatial distribution. It is widely used in the early diagnosis of cancer and other diseases. However, imaging is limited by spontaneous high imaging background, poor penetration depth, and low stability. By using M13 as a multifunctional scaffold, several cell imaging probes aiming at solving these issues were constructed for elevated performance in the fluorescence imaging, magnetic resonance imaging, and near-infrared fluorescence imaging.
Fluorescence imaging
Fluorescence imaging is the most widely used imaging technique among others. M13 as imaging probes has several advantages over conventional fluorophore-labeled antibodies, i.e., nanoscale size, high solubility, multivalency, and orthogonal display. The multivalency facilitates the multiple presentation of both targeting agents and signaling agents, whereas the orthogonal display feature guarantees the two types of agents are functionalized without interfere. Wang’s group [33] simultaneously modified the pVIII protein of M13 phage with fluorescent dye and targeting motif through selective modification of two different reactive groups for cancer cell imaging. The dual-modified M13 phage showed an obviously higher fluorescent signal than that of small molecule fluorescent probe.
Although M13-fluorophore complex was highly specific, the low absorption cress-section and moderate quantum yields of fluorophore resulted in poor sensitivity. Angela M. Belcher’s group proposed a method to enhance the fluorescence by taking advantage of the plasmon-enhanced fluorescence. As shown in Fig. 7A, the pVIII protein of E3 displayed M13 was co-assembled by Cy3 and AgNPs (E3-Cy3-AgNPs complex), producing a 24-fold fluorescence enhancement. A spacer was inserted between pVIII and AgNPs to adjust the distance between Cy3 and AuNPs as their distance is crucial for fluorescence enhancement. The modification of M13 with molecular precision provided a facile platform for evaluating the effect of dye density on the fluorescence enhancement [99]. This M13-based probe was further used for in vitro bacteria detection.
Magnetic resonance imaging (MRI)
Magnetic resonance imaging is a noninvasive cell imaging technique for detecting disease progression. Angela M Belcher’s group demonstrated the M13 phages can be used as scaffold for MRI-tag-modification to enhance imaging selectivity. They displayed poly-glutamatergic peptide motifs on pVIII protein to link the positively charged magnetic nanoparticles based on electrostatic interaction for enhanced light–dark contrast magnetic resonance imaging, and displayed pIII protein with SPARC-binding peptide for tumor targeting. Compared with SPARC-binding peptide functionalized magnetic nanoparticles, the contrast of this approach was enhanced due to the multivalent delivery of contrast agents by M13 [17].
Xe is an attractive MRI contrast agent due to the hyperpolarization property, low toxicity and chemical inertia. Alex Pines’ group modified pVIII with cryptophane-A (CryA) for Xe binding through an N-terminal specific bioconjugation method. About 1050 CryA copies were loaded on each M13, which improved the sensitivity of M13-CryA to 230 fM [100]. By further displaying scFvs against the EGFR on pIII for targeting, molecular targeting against EFGR-positive cancer cells was achieved [101].
Near Infrared (NIR) imaging
Compared with the high background, photobleaching, and low stability characteristics of fluorescence imaging, the NIR possesses the advantages of deep tissue penetration, low autofluorescence, and reduced scattering. Angela M. Belcher’s group assembled single-walled carbon nanotubes (SWNTs) onto the surface of multifunctional M13 phage for the cancer cell NIR imaging. The prepared M13-SWNTs can be detected even at a low dosage of 2 μg/mL and up to 2.5 cm in tissue-like phantoms. Meanwhile, the uptake efficiency of the specific M13-SWNTs by the prostate cancer cell was improved by 4 times than nontargeted ones [102]. Subsequently, the same group further used this system to target ovarian tumor to the depth of 9.7–19.2 mm using reflectance imaging system [100]. This system could also be used for real-time image-guided surgery with a limited pixel resolution of 200 μm [103]. By replacing tumor-targeting peptide with antibodies against bacteria, NIR imaging of pathogenic infections can also be realized. Compared with conventional dyes, SWNT imaging had about 1.4 times more signal amplification and provided 5.7-fold enhancement in the imaging of staphylococcus infection (Fig. 7B) [104].
A Illustration of the synthesis of E3-Cy3-AgNPs complex (upper) and the super resolution fluorescence images of E. coli stained with the E3-CTPEG2K-Cy3-AgNPs probe (below). Scale bar is 1 μm. Copyright 2019 Wiley–VCH GmbH [99]. B Structure of SWNT-based M13 phage (upper) and the imaging examples of E. coli-infected mice with SWNT probe (below). Copyright 2014 Springer Nature [104]
Remaining issues and future perspectives
Albeit great success has been achieved in the application of M13 phages in the field of analytical application, some issues still exist.
First, the specific affinity towards the target by M13 phage is the key to realize highly selective analysis. However, the binding affinity (Kd) of the mono-peptide screened from the phage display library is usually in the level of 1 × 10−6 M, which is far from satisfactory to achieve highly selective analysis. Several possible ways might be useful to improve the binding affinity. (1) Developing more effective peptide libraries, such as cyclic peptide library which possesses more complex spatial configuration thus has better interaction with the target. (2) As the lengthy natural evolution is no doubt more powerful than artificial evolution from limited libraries with only several rounds of biopanning, naturally existing phages possess stringent specificity towards its host bacteria. Therefore, the construction of secondary peptide libraries from naturally existing receptor binding proteins (RPBs) may also provide more opportunities to get phages with higher affinity towards bacteria. (3) Improve the way of biopanning. Currently, in a standard biopanning procedure, phages that bind with the target were stripped with eluates and regarded as the one that bind tightly with the target. But actually those cannot be eluted may have higher affinity towards the target, but was lost during the biopanning process. Besides, current biopanning approach is not capable of selecting phages that bind to a particular site of the target molecule. The way how the target molecule is immobilized during biopanning also determines which parts of the target interact with the phages and what kind of phages is finally selected. All these issues needs systematic exploration. Second, the existing studies mainly used M13 phages as a more effective alternative for signaling molecule-labeled antibody. Researchers always focused on the chemical properties of M13, but omitted its physical/mechanical properties. Our recent study revealed that the flexibility property of M13 facilitated enhanced multivalent interaction between bacteria cells and aptamer-decorated M13 nanofibers. Besides, the swaying motion of this long and flexible nanofiber can also accelerate the mass transport around them, thus improving the sensitivity of our bacteria sensing system. However, it is worth noting that the current length-diameter ratio and stiffness of native M13 may not be the optimal case, where entanglement between M13 nanofibers may occur. A recent study by Belcher’s group manipulated the length of M13 to change in the range of 50 ~ 1300 nm [22]. This opens possibilities to assemble M13 nanofiber array with different length and stiffness, which might be useful in mimicking the extracellular matrix (ECM) and in studying the effect of machinal cues on cell behaviors.
Third, there are still rooms to improve for achieving accurate functionalization of M13 phages at the molecular precision. This is particular important, as studying the interaction between cells or between receptors and ligands on one cell always need analysis tools with molecular precision. However, as chemical modification is chemoselective, the reaction usually takes place in all available reactive sites instead of desired regions or coat proteins. The incorporation of ncAAs with biorthogonal reactive sites onto the surface of M13 can realize site-specific chemical modification, but the currently available ncAA arsenal is still limited. The direct insertion of foreign genes facilitates 100% display of the endogenous peptide on M13 surface, but only within short peptide. With the assistance of transpeptidase, for instance, Sortase A, the size of peptide/protein displayed on M13 surface can be facilely expanded [25, 105, 106]. However, Sortase A-based displaying strategy faced another issue of reaction reversibility [107]. Efforts are still needed to achieve functionalized M13 with definite molecular composition.
In general, although several issues remain to be solved, this field is still in the ascendant. This requires the joint effort from researches in multiple research field including biology, chemistry and material science. As modern analytical science now becomes a multidiscipline of more than just quantification, novel strategies to measure weak interactions, sense the invisibles and monitor the dynamic process are required, where the versatile M13 can also make contributions in the near future.
References
Smith GP, Petrenko VA. Phage Display. Chem Rev. 1997;97:391–410.
Smith GP. Phage Display: Simple Evolution in a Petri Dish (Nobel Lecture). Angew Chem Int Ed. 2019;58(41):14428–37.
Sunderland KS, Yang M, Mao C. Phage-Enabled Nanomedicine: From Probes to Therapeutics in Precision Medicine. Angew Chem Int Ed. 2017;56(8):1964–92.
Aliakbar Ahovan Z, Hashemi A, De Plano LM, Gholipourmalekabadi M, Seifalian A. Bacteriophage Based Biosensors: Trends, Outcomes and Challenges. Nanomaterials-Basel. 2020;10(3):501.
Yue H, Li Y, Yang M, Mao C. T7 Phage as an Emerging Nanobiomaterial with Genetically Tunable Target Specificity. Adv Sci. 2022;9(4):2103645.
Rondot Susanne, Koch Joachim, Breitling F, Dübel S. A helper phage to improve single-chain antibody presentation in phage display. Nat Biotechnol. 2002;19(1):75–8.
Lubkowski J, Hennecke F, Plückthun A, Wlodawer A. The structural basis of phage display elucidated by the crystal structure of the N-terminal domains of g3p. Nat Struct Mol Biol. 1998;5(2):140–7.
Loset GA, Sandlie I. Next generation phage display by use of pVII and pIX as display scaffolds. Methods. 2012;58(1):40–6.
Tornetta M, Reddy R, Wheeler JC. Selection and maturation of antibodies by phage display through fusion to pIX. Methods. 2012;58(1):34–9.
Loset GA, Bogen B, Sandlie I. Expanding the versatility of phage display I: efficient display of peptide-tags on protein VII of the filamentous phage. PLoS ONE. 2011;6(2):e14702.
Haigh NG, Webster RE. The pI and pXI assembly proteins serve separate and essential roles in filamentous phage assembly. J Mol Biol. 1999;293(5):1017–27.
Russel M, Lowman HB, Clackson T. 2004 Introduction to phage biology and phage display. HB Lowman, T Clackson (Eds), Phage Display: A Practical Approach. 266:1–26.
Sidhu SS, Geyer CR. Phage display in biotechnology and drug discovery: CRC Press; 2015.
JoachimMessing. 1991 Cloning in M13 phage or how to use biology at its best. Gene. 100:3–12.
Lee JH, Warner CM, Jin HE, Barnes E, Poda AR, Perkins EJ, Lee SW. Production of tunable nanomaterials using hierarchically assembled bacteriophages. Nat Protoc. 2017;12(9):1999–2013.
Nam KT, Kim DW, Yoo PJ, Chiang CY, Meethong N, Hammond PT, Chiang YM, Belcher AM. Virus-enabled synthesis and assembly of nanowires for lithium ion battery electrodes. Science. 2006;312(5775):885–8.
Ghosh D, Lee Y, Thomas S, Kohli AG, Yun DS, Belcher AM, Kelly KA. M13-templated magnetic nanoparticles for targeted in vivo imaging of prostate cancer. Nat Nanotechnol. 2012;7(10):677–82.
Praetorius F, Kick B, Behler KL, Honemann MN, Weuster-Botz D, Dietz H. Biotechnological mass production of DNA origami. Nature. 2017;552(7683):84–7.
Zhang Q, Xia K, Jiang M, Li Q, Chen W, Han M, Li W, Ke R, Wang F, Zhao Y, Liu Y, Fan C, Gu H. Catalytic DNA-Assisted Mass Production of Arbitrary Single-Stranded DNA. Angew Chem Int Ed. 2023;62(5): e202212011.
Fiskin E, Lareau CA, Ludwig LS, Eraslan G, Liu F, Ring AM, Xavier RJ, Regev A. Single-cell profiling of proteins and chromatin accessibility using PHAGE-ATAC. Nat Biotechnol. 2022;40(3):374–81.
Cha TG, Tsedev U, Ransil A, Embree A, Gordon DB, Belcher AM, Voigt CA. 2021 Genetic Control of Aerogel and Nanofoam Properties, Applied to Ni–MnOx Cathode Design. Adv Funct Mater. 31(35).
Tsedev U, Lin CW, Hess GT, Sarkaria JN, Lam FC, Belcher AM. Phage Particles of Controlled Length and Genome for In Vivo Targeted Glioblastoma Imaging and Therapeutic Delivery. ACS Nano. 2022;16(8):11676–91.
Ledsgaard L, Kilstrup M, Karatt-Vellatt A, McCafferty J, Laustsen AH. Basics of Antibody Phage Display Technology. Toxins (Basel). 2018;10(6):236.
Ulfo L, Cantelli A, Petrosino A, Costantini PE, Nigro M, Starinieri F, Turrini E, Zadran SK, Zuccheri G, Saporetti R, Di Giosia M, Danielli A, Calvaresi M. Orthogonal nanoarchitectonics of M13 phage for receptor targeted anticancer photodynamic therapy. Nanoscale. 2022;14(3):632–41.
Chung WJ, Lee DY, Yoo SY. Chemical modulation of M13 bacteriophage and its functional opportunities for nanomedicine. Int J Nanomedicine. 2014;9:5825–36.
Fang H, Zhan S, Feng L, Chen X, Guo Q, Guo Y, He Q, Xiong Y. Chemical modification of M13 bacteriophage as nanozyme container for dramatically enhanced sensitivity of colorimetric immunosensor. Sens Actuators B. 2021;346:130368.
Mariana Alarcón-Correa J-PG, Troll Jonas, Kadiri Vincent Mauricio, Bill Joachim, Fischer Peer, Rothenstein Dirk. Self-assembled phage-based colloids for high localized enzymatic activity. ACS Nano. 2019;13(5):5810–5.
Yacoby I, Bar H, Benhar I. Targeted drug-carrying bacteriophages as antibacterial nanomedicines. Antimicrob Agents Chemother. 2007;51(6):2156–63.
Bar H, Yacoby I, Benhar I. Killing cancer cells by targeted drug-carrying phage nanomedicines. BMC Biotechnol. 2008;8(1):37.
Benhar LVaI. 2011 In vivo characteristics of targeted drug-carrying filamentous bacteriophage nanomedicines. J Nanobiotechnol. 9(1):58.
Lee JH, Cha JN. Amplified protein detection through visible plasmon shifts in gold nanocrystal solutions from bacteriophage platforms. Anal Chem. 2011;83(9):3516–9.
Murugesan M, Abbineni G, Nimmo SL, Cao B, Mao C. Virus-based photo-responsive nanowires formed by linking site-directed mutagenesis and chemical reaction. Sci Rep. 2013;3:1820.
Li Kai, Chen Yi, Li Siqi, Nguyen HG, Niu Zhongwei, You Shaojin, Mello Charlene M, Xiaobing Lu, Wang Q. Chemical Modification of M13 Bacteriophage and Its Application in Cancer Cell Imaging. Bioconjugate Chem. 2010;21(7):1369–77.
Santoso B, Lam S, Murray BW, Chen G. A simple and efficient maleimide-based approach for peptide extension with a cysteine-containing peptide phage library. Bioorg Med Chem Lett. 2013;23(20):5680–3.
Ng S, Jafari MR, Matochko WL, Derda R. Quantitative synthesis of genetically encoded glycopeptide libraries displayed on M13 phage. ACS Chem Biol. 2012;7(9):1482–7.
Carmody CM, Goddard JM, Nugen SR. Bacteriophage Capsid Modification by Genetic and Chemical Methods. Bioconjugate Chem. 2021;32(3):466–81.
Allen GL, Grahn AK, Kourentzi K, Willson RC, Waldrop S, Guo J, Kay BK. Expanding the chemical diversity of M13 bacteriophage. Front Microbiol. 2022;13:961093.
Karen E. Sandman JSB, and Christopher J. 2000 Noren. Phage Display of Selenopeptides. J Am Chem Soc. 122(5):960–1.
Beech J, Saleh L, Frentzel J, Figler H, Correa IR Jr, Baker B, Ramspacher C, Marshall M, Dasa S, Linden J, Noren CJ, Kelly KA. Multivalent site-specific phage modification enhances the binding affinity of receptor ligands. Bioconjugate Chem. 2015;26(3):529–36.
Tian F, Tsao M-L, Schultz PG. A Phage Display System with Unnatural Amino Acids. J Am Chem Soc. 2004;126(49):15962–3.
Meng-Lin Tsao Dr. FTD, Peter G. Schultz. 2005 Selective Staudinger Modification of Proteins Containing p-Azidophenylalanine. ChemBioChem. 6(12):2147–9.
Liu CC, Mack AV, Brustad EM, Mills JH, Groff D, Smider VV, Schultz PG. Evolution of Proteins with Genetically Encoded “Chemical Warheads.” J Am Chem Soc. 2009;131(28):9616–7.
Chang C, Liu AVM, Tsao Meng-Lin, Mills Jeremy H, Lee Hyun Soo, Choe Hyeryun, Farzan Michael, Schultz Peter G, Smider Vaughn V. Protein evolution with an expanded genetic code. Proc Natl Acad Sci U S A. 2008;105(46):17688–93.
Mingchao Kang KL, Ai Hui-wang, Shen Weijun, Kim Chan Hyuk, Chen Peng R, Lee Hyun Soo, Solomon Edward I, Schultz Peter G. Evolution of Iron(II)-Finger Peptides by Using a Bipyridyl Amino Acid. ChemBioChem. 2014;15(6):822–5.
Oller-Salvia B, Chin JW. Efficient Phage Display with Multiple Distinct Non-Canonical Amino Acids Using Orthogonal Ribosome-Mediated Genetic Code Expansion. Angew Chem Int Ed. 2019;58(32):10844–8.
Wang XS, Chen PC, Hampton JT, Tharp JM, Reed CA, Das SK, Wang DS, Hayatshahi HS, Shen Y, Liu J, Liu WR. A Genetically Encoded, Phage-Displayed Cyclic-Peptide Library. Angew Chem Int Ed. 2019;58(44):15904–9.
Xu P, Ghosh S, Gul AR, Bhamore JR, Park JP, Park TJ. Screening of specific binding peptides using phage-display techniques and their biosensing applications. TrAC Trends Anal Chem. 2021;137:116229.
Smith GP, Petrenko VA. Phage Display. Chem Rev. 1997;97(2):391–410.
Peltomaa R, Benito-Peña E, Barderas R, Moreno-Bondi MC. Phage Display in the Quest for New Selective Recognition Elements for Biosensors. ACS Omega. 2019;4(7):11569–80.
Li Y, Qu X, Cao B, Yang T, Bao Q, Yue H, Zhang L, Zhang G, Wang L, Qiu P, Zhou N, Yang M, Mao C. Selectively Suppressing Tumor Angiogenesis for Targeted Breast Cancer Therapy by Genetically Engineered Phage. Adv Mater. 2020;32(29):e2001260.
Sellers DL, Tan JY, Pineda JMB, Peeler DJ, Porubsky VL, Olden BR, Salipante SJ, Pun SH. Targeting Ligands Deliver Model Drug Cargo into the Central Nervous System along Autonomic Neurons. ACS Nano. 2019;13(10):10961–71.
Wang J, Tan Y, Ling J, Zhang M, Li L, Liu W, Huang M, Song J, Li A, Song Y, Yang C, Zhu Z. Highly paralleled emulsion droplets for efficient isolation, amplification, and screening of cancer biomarker binding phages. Lab Chip. 2021;21(6):1175–84.
Junxia Wang LL. Yingkun Zhang, Kaifeng Zhao, Xiaofeng Chen, Haicong Shen, Yuanqiang Chen, Jia Song, Yuqiang Ma, Chaoyong Yang, Hongming Ding, and Zhi Zhu Synergetic collision and space separation in microfluidic chip for efficient affinity-discriminated molecular selection. Proc Natl Acad Sci U S A. 2022;119(41):e2211538119.
Philpott DN, Gomis S, Wang H, Atwal R, Kelil A, Sack T, Morningstar B, Burnie C, Sargent EH, Angers S, Sidhu S, Kelley SO. Rapid On-Cell Selection of High-Performance Human Antibodies. ACS Cent Sci. 2022;8(1):102–9.
Muzard J, Platt M, Lee GU. M13 bacteriophage-activated superparamagnetic beads for affinity separation. Small. 2012;8(15):2403–11.
Jeon CS, Hwang I, Chung TD. Virus-Tethered Magnetic Gold Microspheres with Biomimetic Architectures for Enhanced Immunoassays. Adv Funct Mater. 2013;23(12):1484–9.
Jo SM, Lee JJ, Heu W, Kim HS. Nanotentacle-structured magnetic particles for efficient capture of circulating tumor cells. Small. 2015;11(16):1975–82.
Yang T, Zhang XY, Zhang XX, Chen ML, Wang JH. Chromium(III) Binding Phage Screening for the Selective Adsorption of Cr(III) and Chromium Speciation. ACS Appl Mater Interfaces. 2015;7(38):21287–94.
Zhu G, Zhao P, Deng N, Tao D, Sun L, Liang Z, Zhang L, Zhang Y. Single chain variable fragment displaying M13 phage library functionalized magnetic microsphere-based protein equalizer for human serum protein analysis. Anal Chem. 2012;84(18):7633–7.
Lang Q, Wang F, Yin L, Liu M, Petrenko VA, Liu A. Specific probe selection from landscape phage display library and its application in enzyme-linked immunosorbent assay of free prostate-specific antigen. Anal Chem. 2014;86(5):2767–74.
You T, Ding Y, Chen H, Song G, Huang L, Wang M, Hua X. Development of competitive and noncompetitive immunoassays for clothianidin with high sensitivity and specificity using phage-displayed peptides. J Hazard Mater. 2022;425:128011.
Germana L, Enza F, Federica C, Laura MDP, Maria P, Domenico F, Marco SN, Santina C, Sebastiano T, Alessandro A, Fortunato N, Caterina M, Salvatore PPG. Phage–AgNPs complex as SERS probe for U937 cell identification. Biosens Bioelectron. 2015;74:398–405.
De Plano LM, Fazio E, Rizzo MG, Franco D, Carnazza S, Trusso S, Neri F, Guglielmino SPP. Phage-based assay for rapid detection of bacterial pathogens in blood by Raman spectroscopy. J Immunol Methods. 2019;465:45–52.
Koh EH, Mun C, Kim C, Park SG, Choi EJ, Kim SH, Dang J, Choo J, Oh JW, Kim DH, Jung HS. M13 Bacteriophage/Silver Nanowire Surface-Enhanced Raman Scattering Sensor for Sensitive and Selective Pesticide Detection. ACS Appl Mater Interfaces. 2018;10(12):10388–97.
Hsieh ST, Cheeney JE, Ding X, Myung NV, Haberer ED. Near-field electrospinning of polymer/phage whispering gallery mode microfiber resonators for label-free biosensing. Sens Actuators B. 2022;367(15):132062.
Lee JH, Domaille DW, Cha JN. Amplified Protein Detection and Identification through DNA-Conjugated M13 Bacteriophage. ACS Nano. 2012;6(6):5621–6.
Domaille DW, Lee JH, Cha JN. High density DNA loading on the M13 bacteriophage provides access to colorimetric and fluorescent protein microarray biosensors. Chem Commun. 2013;49(17):1759–61.
Lee JH, Xu PF, Domaille DW, Choi C, Jin S, Cha JN. M13 Bacteriophage as Materials for Amplified Surface Enhanced Raman Scattering Protein Sensing. Adv Funct Mater. 2014;24(14):2079–84.
Wang XY, Yang JY, Wang YT, Zhang HC, Chen ML, Yang T, Wang JH. M13 phage-based nanoprobe for SERS detection and inactivation of Staphylococcus aureus. Talanta. 2021;221:121668.
Liu J, Ma P, Yu H, Wang M, Yin P, Pang S, Jiao Y, Dong T, Liu A. Discovery of a Phage Peptide Specifically Binding to the SARS-CoV-2 Spike S1 Protein for the Sensitive Phage-Based Enzyme-Linked Chemiluminescence Immunoassay of the SARS-CoV-2 Antigen. Anal Chem. 2022;94(33):11591–9.
Peng H, Chen IA. Rapid Colorimetric Detection of Bacterial Species through the Capture of Gold Nanoparticles by Chimeric Phages. ACS Nano. 2019;13(2):1244–52.
Wu L, Huang T, Yang L, Pan J, Zhu S, Yan X. Sensitive and selective bacterial detection using tetracysteine-tagged phages in conjunction with biarsenical dye. Angew Chem Int Ed. 2011;50(26):5873–7.
Wu L, Luan T, Yang X, Wang S, Zheng Y, Huang T, Zhu S, Yan X. Trace detection of specific viable bacteria using tetracysteine-tagged bacteriophages. Anal Chem. 2014;86(1):907–12.
Wang Y, Li P, Majkova Z, Bever CR, Kim HJ, Zhang Q, Dechant JE, Gee SJ, Hammock BD. Isolation of alpaca anti-idiotypic heavy-chain single-domain antibody for the aflatoxin immunoassay. Anal Chem. 2013;85(17):8298–303.
Lei J, Li P, Zhang Q, Wang Y, Zhang Z, Ding X, Zang W. Anti-idiotypic nanobody-phage based real-time immuno-PCR for detection of hepatocarcinogen aflatoxin in grains and feedstuffs. Anal Chem. 2014;86(21):10841–6.
Brasino MD, Cha JN. Isothermal rolling circle amplification of virus genomes for rapid antigen detection and typing. Analyst. 2015;140(15):5138–44.
Hua X, Yin W, Shi H, Li M, Wang Y, Wang H, Ye Y, Kim HJ, Gee SJ, Wang M, Liu F, Hammock BD. Development of phage immuno-loop-mediated isothermal amplification assays for organophosphorus pesticides in agro-products. Anal Chem. 2014;86(16):8441–7.
Pollock SB, Hu A, Mou Y, Martinko AJ, Julien O, Hornsby M, Ploder L, Adams JJ, Geng H, Muschen M, Sidhu SS, Moffat J, Wells JA. Highly multiplexed and quantitative cell-surface protein profiling using genetically barcoded antibodies. Proc Natl Acad Sci U S A. 2018;115(11):2836–41.
Ma Y, Chen K, Xia F, Atwal R, Wang H, Ahmed SU, Cardarelli L, Lui I, Duong B, Wang Z, Wells JA, Sidhu SS, Kelley SO. Phage-Based Profiling of Rare Single Cells Using Nanoparticle-Directed Capture. ACS Nano. 2021;15(12):19202–10.
Oh JW, Chung WJ, Heo K, Jin HE, Lee BY, Wang E, Zueger C, Wong W, Meyer J, Kim C, Lee SY, Kim WG, Zemla M, Auer M, Hexemer A, Lee SW. Biomimetic virus-based colourimetric sensors. Nat Commun. 2014;5:3043.
Lee JH, Fan B, Samdin TD, Monteiro DA, Desai MS, Scheideler O, Jin HE, Kim S, Lee SW. Phage-Based Structural Color Sensors and Their Pattern Recognition Sensing System. ACS Nano. 2017;11(4):3632–41.
Moon JS, Lee Y, Shin DM, Kim C, Kim WG, Park M, Han J, Song H, Kim K, Oh JW. Identification of Endocrine Disrupting Chemicals using a Virus-Based Colorimetric Sensor. Chem Asian J. 2016;11(21):3097–101.
Moon JS, Kim WG, Shin DM, Lee SY, Kim C, Lee Y, Han J, Kim K, Yoo SY, Oh JW. Bioinspired M-13 bacteriophage-based photonic nose for differential cell recognition. Chem Sci. 2017;8(2):921–7.
Moon J-S, Park M, Kim W-G, Kim C, Hwang J, Seol D, Kim C-S, Sohn J-R, Chung H, Oh J-W. M-13 bacteriophage based structural color sensor for detecting antibiotics. Sens Actuators B. 2017;240:757–62.
Wang X, Yang T, Zhang X, Chen M, Wang J. In situ growth of gold nanoparticles on Hg(2+)-binding M13 phages for mercury sensing. Nanoscale. 2017;9(43):16728–34.
Zeng Y, Yue H, Cao B, Li Y, Yang M, Mao C. Target-Triggered Formation of Artificial Enzymes on Filamentous Phage for Ultrasensitive Direct Detection of Circulating miRNA Biomarkers in Clinical Samples. Angew Chem Int Ed. 2022;61(45):e202210121.
Zhou J, Qi Q, Wang C, Qian Y, Liu G, Wang Y, Fu L. Surface plasmon resonance (SPR) biosensors for food allergen detection in food matrices. Biosens Bioelectron. 2019;142:111449.
Karoonuthaisiri N, Charlermroj R, Morton MJ, Oplatowska-Stachowiak M, Grant IR, Elliott CT. Development of a M13 bacteriophage-based SPR detection using Salmonella as a case study. Sens Actuators B. 2014;190:214–20.
Yoo SY, Oh JW, Lee SW. Phage-chips for novel optically readable tissue engineering assays. Langmuir. 2012;28(4):2166–72.
Kim WG, Song H, Kim C, Moon JS, Kim K, Lee SW, Oh JW. Biomimetic self-templating optical structures fabricated by genetically engineered M13 bacteriophage. Biosens Bioelectron. 2016;85:853–9.
Jin H, Won N, Ahn B, Kwag J, Heo K, Oh JW, Sun Y, Cho SG, Lee SW, Kim S. Quantum dot-engineered M13 virus layer-by-layer composite films for highly selective and sensitive turn-on TNT sensors. Chem Commun. 2013;49(54):6045–7.
Chen L, Wu Y, Lin Y, Wang Q. Virus-templated FRET platform for the rational design of ratiometric fluorescent nanosensors. Chem Commun. 2015;51(50):10190–3.
Kim I, Song H, Kim C, Kim M, Kyhm K, Kim K, Oh JW. Intermolecular distance measurement with TNT suppressor on the M13 bacteriophage-based Forster resonance energy transfer system. Sci Rep. 2019;9(1):496.
Cao Y, Wu N, Li H, Xue J-W, Wang R, Yang T, Wang J-H. Efficient Pathogen Capture and Sensing Promoted by Dynamic Deformable Nanointerfaces. Small. 2022;18(51):2203962.
Sedki M, Chen X, Chen C, Ge X, Mulchandani A. Non-lytic M13 phage-based highly sensitive impedimetric cytosensor for detection of coliforms. Biosens Bioelectron. 2020;148:111794.
Han L, Wang D, Yan L, Petrenko VA, Liu A. Specific phages-based electrochemical impedimetric immunosensors for label-free and ultrasensitive detection of dual prostate-specific antigens. Sens Actuators B. 2019;297:126727.
Shin JH, Park TJ, Hyun MS, Park JP. A phage virus-based electrochemical biosensor for highly sensitive detection of ovomucoid. Food Chem. 2022;378:132061.
Shi R, Zou W, Zhao Z, Wang G, Guo M, Ai S, Zhou Q, Zhao F, Yang Z. Development of a sensitive phage-mimotope and horseradish peroxidase based electrochemical immunosensor for detection of O. O-dimethyl organophosphorus pesticides Biosens Bioelectron. 2022;218:114748.
Huang S, Qi J, deQuilettes DW, Huang M, Lin CW, Bardhan NM, Dang X, Bulovic V, Belcher AM. M13 Virus-Based Framework for High Fluorescence Enhancement. Small. 2019;15(28):e1901233.
Ghosh D, Bagley AF, Na YJ, Birrer MJ, Bhatia SN, Belcher AM. Deep, noninvasive imaging and surgical guidance of submillimeter tumors using targeted M13-stabilized single-walled carbon nanotubes. Proc Natl Acad Sci U S A. 2014;111(38):13948–53.
Palaniappan KK, Ramirez RM, Bajaj VS, Wemmer DE, Pines A, Francis MB. Molecular imaging of cancer cells using a bacteriophage-based 129Xe NMR biosensor. Angew Chem Int Ed. 2013;52(18):4849–53.
Yi H, Ghosh D, Ham MH, Qi J, Barone PW, Strano MS, Belcher AM. M13 phage-functionalized single-walled carbon nanotubes as nanoprobes for second near-infrared window fluorescence imaging of targeted tumors. Nano Lett. 2012;12(3):1176–83.
Ceppi L, Bardhan NM, Na Y, Siegel A, Rajan N, Fruscio R, Del Carmen MG, Belcher AM, Birrer MJ. Real-Time Single-Walled Carbon Nanotube-Based Fluorescence Imaging Improves Survival after Debulking Surgery in an Ovarian Cancer Model. ACS Nano. 2019;13(5):5356–65.
Bardhan NM, Ghosh D, Belcher AM. Carbon nanotubes as in vivo bacterial probes. Nat Commun. 2014;5:4918.
Hess GT, Cragnolini JJ, Popp MW, Allen MA, Dougan SK, Spooner E, Ploegh HL, Belcher AM, Guimaraes CP. M13 bacteriophage display framework that allows sortase-mediated modification of surface-accessible phage proteins. Bioconjugate Chem. 2012;23(7):1478–87.
Ding Y, Chen H, Li J, Huang L, Song G, Li Z, Hua X, Gonzalez-Sapienza G, Hammock BD, Wang M. Sortase-Mediated Phage Decoration for Analytical Applications. Anal Chem. 2021;93(34):11800–8.
Morgan HE, Turnbull WB, Webb ME. Challenges in the use of sortase and other peptide ligases for site-specific protein modification. Chem Soc Rev. 2022;51(10):4121–45.
Funding
This work received financial support from the Natural Science Foundation of China (22074011, 21874014, 21727811), the Fundamental Research Funds for the Central Universities (N2005017, N2005027) and the Liaoning Revitalization Talents Program (XLYC2007102, XLYC1802016).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Published in the topical collection Young Investigators in (Bio-)Analytical Chemistry 2023 with guest editors Zhi-Yuan Gu, Beatriz Jurado-Sánchez, Thomas H. Linz, Leandro Wang Hantao, Nongnoot Wongkaew, and Peng Wu.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, R., Li, HD., Cao, Y. et al. M13 phage: a versatile building block for a highly specific analysis platform. Anal Bioanal Chem 415, 3927–3944 (2023). https://doi.org/10.1007/s00216-023-04606-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-023-04606-w