Abstract
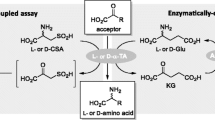
ω-Transaminases (ω-TAs) are widely available for the production of chiral amines and unnatural amino acids. Herein, a rapid spectrophotometric method was developed for screening ω-TAs based on the colored products that can be generated from transamination reactions between aliphatic α-diketones and amino donors catalyzed by ω-TAs. The possible mechanism of the formation of the colored product was investigated according to LC-Q-TOF–MS analysis. Among seven diketones, 2,3-butanedione was selected as the most suitable amino acceptor for colorimetric screening of ω-TAs with high efficiency, high sensitivity, and low background interference. Meanwhile, the absorbance of the colored product generated by 2,3-butanedione catalyzed by ω-TAs in this method was linearly correlated with the results by HPLC analysis. This method was also confirmed to effectively screen ω-TA mutants with high activity towards isopropylamine.
Graphical Abstract










Similar content being viewed by others
Data availability
All proteins in the present study are available from GenBank via the accession codes. The original data are provided with the paper.
References
Eliot AC, Kirsch JF. Pyridoxal phosphate enzymes: mechanistic, structural, and evolutionary considerations. Annu Rev Biochem. 2004;73(1):383–415. https://doi.org/10.1146/annurev.biochem.73.011303.074021.
Han SW, Kim J, Cho HS, Shin JS. Active site engineering of ω-transaminase guided by docking orientation analysis and virtual activity screening. ACS Catal. 2017;7(6):3752–62. https://doi.org/10.1021/acscatal.6b03242.
Mathew S, Yun H. ω-Transaminases for the production of optically pure amines and unnatural amino acids. ACS Catal. 2012;2(6):993–1001. https://doi.org/10.1016/j.jbiotec.2015.01.011.
Slabu I, Galman JL, Lloyd RC, Turner NJ. Discovery, engineering and synthetic application of transaminase biocatalysts. ACS Catal. 2017;7(12):8263–84. https://doi.org/10.1021/acscatal.7b02686.
Savile CK, Janey JM, Mundorff EC, Moore JC, Tarn S, Jarvis WR, Colbeck JC, Krebber A, Fleitz FJ, Brands J. Biocatalytic asymmetric synthesis of chiral amines from ketones applied to sitagliptin manufacture. Science. 2010;329(5989):305–9. https://doi.org/10.1126/science.1188934.
Novick SJ, Dellas N, Garcia R, Ching C, Bautista A, Homan D, Alvizo O, Entwistle D, Kleinbeck F, Schlama T, Ruch T. Engineering an amine transaminase for the efficient production of a chiral sacubitril precursor. ACS Catal. 2021;11:3762–70. https://doi.org/10.1021/acscatal.0c05450.
Kelly SA, Mix S, Moody TS, Gilmore BF. Transaminases for industrial biocatalysis: novel enzyme discovery. Appl Microbiol Biotechnol. 2020;104:4781–94. https://doi.org/10.1007/s00253-020-10585-0.
Lutz S, Iamurri SM. Protein engineering: past, present, and future. Methods Mol Biol. 2018;1685:1–12. https://doi.org/10.1007/978-1-4939-7366-8_1.
Mathew S, Shin G, Shon M, Yun H. High throughput screening methods for ω-transaminases. Biotechnol Bioproc E. 2013;18(1):1–7. https://doi.org/10.1007/s12257-012-0544-x.
Hwang BY, Kim BG. High-throughput screening method for the identification of active and enantioselective ω-transaminases. Enzyme Microb Tech. 2004;34(5):429–36. https://doi.org/10.1016/j.enzmictec.2003.11.019.
Hopwood J, Truppo MD, Turner NJ, Lloyd RC. A fast and sensitive assay for measuring the activity and enantioselectivity of transaminases. Chem Commun. 2011;47(2):773–5. https://doi.org/10.1039/c0cc02919j.
Green AP, Turner NJ, Reilly OE. Chiral amine synthesis using ω-transaminases: an amine donor that displaces equilibria and enables high-throughput screening. Angew Chem Int Ed Engl. 2014;53(40):10714–7. https://doi.org/10.1002/anie.201406571.
Baud D, Ladkau N, Moody TS, Ward JM, Hailes HC. A rapid, sensitive colorimetric assay for the high-throughput screening of transaminases in liquid or solid-phase. Chem Commun. 2015;51(97):17225–8. https://doi.org/10.1039/c5cc06817g.
Cheng F, Chen XL, Xiang C, Liu ZQ, Zheng YG. Fluorescence-based high-throughput screening system for R-ω-transaminase engineering and its substrate scope extension. Appl Microbiol Biot. 2020;104(7):1–11. https://doi.org/10.1007/s00253-020-10444-y.
Shin JS, Yun H, Jang JW, Park I, Kim BG. Purification, characterization, and molecular cloning of a novel amine: pyruvate transaminase from Vibrio fluvialis JS17. Appl Microbiol Biotechnol. 2003;61(5–6):463–71. https://doi.org/10.1007/s00253-003-1250-6.
Park E, Kim M, Shin JS. One-pot conversion of L-threonine into L-homoalanine: biocatalytic production of an unnatural amino acid from a natural one. Adv Synth Catal. 2010;352:3391–8. https://doi.org/10.1002/adsc.201000601.
Tang KX, Yi YF, Gao Z, Jia HH, Li Y, Cao F, Jiang M, Wei P. Identification, heterologous expression and characterization of a transaminase from Rhizobium sp. Catal Lett. 2020;150:2415–26. https://doi.org/10.1007/s10562-020-03121-2.
Guan LJ, Ohtsuka J, Okai M, Miyakawa T, Mase T, Zhi Y, Hou F, Ito N, Iwasaki A, Yasohara Y, Tanokura M. A new target region for changing the substrate specificity of amine transaminases. Sci Rep. 2015;5:10753. https://doi.org/10.1038/srep10753.
Nierman WC, Pain A, Anderson MJ, et al. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature. 2005;438(7071):1151–6. https://doi.org/10.1038/nature04332.
Jiang J, Chen X, Zhang D, Wu Q, Zhu D. Characterization of (R)-selective amine transaminases identified by in silico motif sequence blast. Appl Microbiol Biotechnol. 2015;99(6):2613–21. https://doi.org/10.1007/s00253-014-6056-1.
Kelly SA, Pohle S, Wharry S, Mix S, Allen C, Moody TS, Gilmore BF. Application of ω-transaminases in the pharmaceutical industry. Chem Rev. 2017;118(1):349–67. https://doi.org/10.1021/acs.chemrev.7b00437.
Iwasaki M, Hayashi H, Kagamiyama H. Protonation state of the active-site Schiff base of aromatic amino acid aminotransferase: modulation by binding of ligands and implications for its role in catalysis. J Biochem. 1994;115(1):156–61. https://doi.org/10.1093/oxfordjournals.jbchem.a124293.
Nobili A, Munsberg FS, Kohls H, Trentin I, Carola Schulzke, Hçhne M, Bornscheuer UT. Engineering the active site of the amine transaminasefrom Vibrio fluvialis for the asymmetric synthesis of aryl–alkyl amines and amino alcohols. ChemCatChem. 2014;7(5):757–60. https://doi.org/10.1002/cctc.201403010.
Willies SC, Galman JL, Slabu I, Turner NJ. Rapid screening and scale-up of transaminase catalysed reactions. Org Biomol Chem. 2009;7(2):395–8. https://doi.org/10.1039/b817730a.
Zhang JD, Wu HL, Meng T, Zhang CF, Fan XJ, Chang HH, Wei WL. A high-throughput microtiter plate assay for the discovery of active and enantioselective amino alcohol-specific transaminases. Anal Bioch. 2017;518:94–101. https://doi.org/10.1016/j.ab.2016.11.015.
Cairns R, Gomm A, Peel C, Sharkey M, Reilly EO. A comprehensive quantitative assay for amine transaminases. ChemCatChem. 2019;11:4738–43. https://doi.org/10.1021/pr301178a.
Stratidakis KP, Ergas TT, Pavlidis IV. The challenge of using isopropylamine as an amine donor in transaminase catalysed reactions. Org Biomol Chem. 2019;17(7):1634–42. https://doi.org/10.1039/c8ob02342e.
Dawood AWH, Weiß MS, Schulz C, Pavlidis IV, Iding H, de Souza ROMA, Bornscheuer UT. Isopropylamine as amine donor in transaminase-catalyzed reactions: better acceptance through reaction and enzyme engineering. ChemCatChem. 2018;10(18):3943–9. https://doi.org/10.1002/cctc.201800936.
Morrison KL, Weiss GA. Combinatorial alanine-scanning. Opin Chem Biol. 2001;5(3):302–7. https://doi.org/10.1016/S1367-5931(00)00206-4.
Cassimjee KE, Humble MS, Miceli V, Colomina CG, Berglund P. Active site quantification of an ω-transaminase by performing a half transamination reaction. ACS Catal. 2011;1(9):1051–5. https://doi.org/10.1021/cs200315h.
Acknowledgements
The authors would like to thank the Jiangsu Synergetic Innovation Center for Advanced Bio-manufacture and PAPD to fund this work. For Feng Dan and Shuai Shi, we also thank their work on language checking. The authors also gratefully acknowledge the editors and the reviewers for their useful feedback which improved this paper.
Funding
This work was financially funded by the Jiangsu Synergetic Innovation Center for Advanced Bio-manufacture and PAPD.
Author information
Authors and Affiliations
Contributions
K.X. Tang conducted and designed most parts of the experiments. J.C. Dong assisted the first author with HPLC analysis. Z.H. Zheng, T. Zhang, and H.Y. Pan performed activity and data processing. Y. Li. and H.H. Jia. designed and supervised the project. P. Wei was the coordinator of the project. All authors discussed the design and results, commented on the manuscript, and approved the manuscript.
Corresponding authors
Ethics declarations
Conflicts of Interest
The authors declare that there is no conflict of interest.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Tang, K., Dong, J., Zheng, Z. et al. The rapid high-throughput screening of ω-transaminases via a colorimetric method using aliphatic α-diketones as amino acceptors. Anal Bioanal Chem 415, 1733–1740 (2023). https://doi.org/10.1007/s00216-023-04573-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-023-04573-2