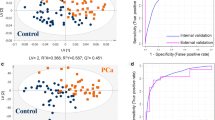
Abstract
Non-Hodgkin’s lymphoma (NHL) is a cancer of the lymphatic system where the lymphoid and hematopoietic tissues are infiltrated by malignant neoplasms of B, T, and natural killer lymphocytes. Effective and less invasive methods for NHL screening are urgently needed. Herein, we report an untargeted gas chromatography-mass spectrometry (GC-MS) method to investigate metabolic changes in non-volatile derivatized compounds from urine samples of NHL patients (N = 15) and compare them to healthy controls (N = 34). Uni- and multivariate data analysis showed 18 endogenous metabolites, including amino acids and their metabolites, sugars, small organic acids, and vitamins, as statistically significant for group differentiation. A receiver operating characteristic curve (ROC) generated from a support vector machine (SVM) algorithm-based model achieved 0.998 of predictive accuracy, displaying the potential and relevance of GC-MS-detected urinary non-volatile compounds for predictive purposes. Furthermore, a specific panel of key metabolites was also evaluated, showing similar results. All in all, our results indicate that this robust GC-MS method is an effective screening tool for NHL diagnosis and it is able to highlight different pathways of the disease.

Graphical Abstract





Similar content being viewed by others
Data availability
The datasets generated during and/or analyzed during the current study are under curation process in the Metabolights repository, https://www.ebi.ac.uk/metabolights/MTBLS1816
References
Pearce L. Non-Hodgkin’s lymphoma. Nurs Stand. 2016;31:15.
Ansell SM. Non-Hodgkin lymphoma: diagnosis and treatment. Mayo Clin Proc. 2015;90:1152–63.
Shankland KR, Armitage JO, Hancock BW. Non-Hodgkin lymphoma. Lancet. 2012;380:848–57.
Non-Hodgkin lymphoma In: Global Cancer Observatory https://gco.iarc.fr/today/data/factsheets/cancers/34-Non-hodgkin-lymphoma-fact-sheet.pdf. Accessed 11 Jan 2020.
Key Statistics for Non-Hodgkin Lymphoma In: American Cancer Society https://www.cancer.org/cancer/non-hodgkin-lymphoma/about/key-statistics.html. Accessed 10 Dec 2019.
Ansell SM, Armitage J. Non-Hodgkin lymphoma: diagnosis and treatment. Mayo Clin Proc. 2005;80:1087–97.
Marchand CR, Farshidfar F, Rattner J, Bathe OF. A framework for development of useful metabolomic biomarkers and their effective knowledge translation. Metabolites. 2018;8:59.
Barichello S, Deng L, Ismond KP, Loomes DE, Kirwin EM, Wang H, et al. Comparative effectiveness and cost-effectiveness analysis of a urine metabolomics test vs. alternative colorectal cancer screening strategies. Int J Color Dis. 2019;34:1953–62.
Patti GJ, Yanes O, Siuzdak G. Innovation: metabolomics: the apogee of the omics trilogy. Nat Rev Mol Cell Biol. 2012;13:263–9.
Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–33.
Kumar A, Misra BB. Challenges and opportunities in cancer metabolomics. Proteomics. 2019;19:e1900042.
Barberini L, Restivo A, Noto A, Deidda S, Fattuoni C, Fanos V, et al. A gas chromatography-mass spectrometry (GC-MS) metabolomic approach in human colorectal cancer (CRC): the emerging role of monosaccharides and amino acids. Ann Transl Med. 2019;7:727.
Chan EC, Koh PK, Mal M, Cheah PY, Eu KW, Backshall A, et al. Metabolic profiling of human colorectal cancer using high-resolution magic angle spinning nuclear magnetic resonance (HR-MAS NMR) spectroscopy and gas chromatography mass spectrometry (GC/MS). J Proteome Res. 2009;8:352–61.
Spivack S, Shi MK, Patel D, Desai A, Dobkin J, Shah C, et al. P1.11-11 initial discovery of exhaled small polar energetics-related metabolites by GC-MS for lung cancer risk assessment. J Thorac Oncol. 2019;14:S519.
Filipiak W, Sponring A, Filipiak A, Ager C, Schubert J, Miekisch W, et al. TD-GC-MS analysis of volatile metabolites of human lung cancer and normal cells in vitro. Cancer Epidemiol Biomark Prev. 2010;19:182–95.
Poli D, Goldoni M, Corradi M, Acampa O, Carbognani P, Internullo E, et al. Determination of aldehydes in exhaled breath of patients with lung cancer by means of on-fiber-derivatisation SPME-GC/MS. J Chromatogr B Analyt Technol Biomed Life Sci. 2010;878:2643–51.
Phillips M, Altorki N, Austin JH, Cameron RB, Cataneo RN, Greenberg J, et al. Prediction of lung cancer using volatile biomarkers in breath. Cancer Biomark. 2007;3:95–109.
Rodrigues D, Pinto J, Araujo AM, Jeronimo C, Henrique R, Bastos ML, et al. GC-MS metabolomics reveals distinct profiles of low- and high-grade bladder cancer cultured cells. Metabolites. 2019;9.
Zhou Y, Song R, Ma C, Zhou L, Liu X, Yin P, et al. Discovery and validation of potential urinary biomarkers for bladder cancer diagnosis using a pseudotargeted GC-MS metabolomics method. Oncotarget. 2017;8:20719–28.
Zhou Y, Song R, Zhang Z, Lu X, Zeng Z, Hu C, et al. The development of plasma pseudotargeted GC-MS metabolic profiling and its application in bladder cancer. Anal Bioanal Chem. 2016;408:6741–9.
Hadi NI, Jamal Q, Iqbal A, Shaikh F, Somroo S, Musharraf SG. Serum metabolomic profiles for breast cancer diagnosis, grading and staging by gas chromatography-mass spectrometry. Sci Rep. 2017;7:1715.
Lima AR, Araujo AM, Pinto J, Jeronimo C, Henrique R, Bastos ML, et al. Discrimination between the human prostate normal and cancer cell exometabolome by GC-MS. Sci Rep. 2018;8:5539.
Lima AR, Araujo AM, Pinto J, Jeronimo C, Henrique R, Bastos ML, et al. GC-MS-based endometabolome analysis differentiates prostate cancer from normal prostate cells. Metabolites. 2018;8.
Wu H, Liu T, Ma C, Xue R, Deng C, Zeng H, et al. GC/MS-based metabolomic approach to validate the role of urinary sarcosine and target biomarkers for human prostate cancer by microwave-assisted derivatization. Anal Bioanal Chem. 2011;401:635–46.
Ibanez C, Simo C, Palazoglu M, Cifuentes A. GC-MS based metabolomics of colon cancer cells using different extraction solvents. Anal Chim Acta. 2017;986:48–56.
Chen Y, Zhang J, Guo L, Liu L, Wen J, Xu L, et al. A characteristic biosignature for discrimination of gastric cancer from healthy population by high throughput GC-MS analysis. Oncotarget. 2016;7:87496–510.
Hu JD, Tang HQ, Zhang Q, Fan J, Hong J, Gu JZ, et al. Prediction of gastric cancer metastasis through urinary metabolomic investigation using GC/MS. World J Gastroenterol. 2011;17:727–34.
Jiang G, Shen X, Kang H, Li K, Zheng J, Yu Y. Serum metabolite profiling of cutaneous T-cell lymphoma based on a multiplatform approach. J Chromatogr B Analyt Technol Biomed Life Sci. 2018;1077–1078:71–6.
Zhou J, Yu S, Wang Y, Gu X, Wu Q, Xue Y, et al. Serum metabolite profiling of B-cell non-Hodgkin’s lymphoma using UPLC-QTOFMS and GC-TOFMS. Metabolomics. 2013;10:677–87.
Hua Q, Wang L, Liu C, Han L, Zhang Y, Liu H. Volatile metabonomic profiling in urine to detect novel biomarkers for B-cell non-Hodgkin’s lymphoma. Oncol Lett. 2018;15:7806–16.
Tsugawa H, Cajka T, Kind T, Ma Y, Higgins B, Ikeda K, et al. MS-DIAL: data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat Methods. 2015;12:523–6.
Sumner LW, Amberg A, Barrett D, Beale MH, Beger R, Daykin CA, et al. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics. 2007;3:211–21.
Chong J, Soufan O, Li C, Caraus I, Li S, Bourque G, et al. MetaboAnalyst 4.0: towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018;46:W486–W94.
Eriksson L, Byrne T, Johansson E, Trygg J, Vikström C. Multi- and megavariate data analysis basic principles and applications. 3rd ed. Umeå: Umetrics Academy; 2013.
Triba MN, Le Moyec L, Amathieu R, Goossens C, Bouchemal N, Nahon P, et al. PLS/OPLS models in metabolomics: the impact of permutation of dataset rows on the K-fold cross-validation quality parameters. Mol BioSyst. 2015;11:13–9.
Moritz T, Johansson A. Plant metabolomics. In: Griffiths W, editor. Metabolomics, metabonomics and metabolite profiling. Cambridge: Royal Society of Chemistry; 2007. p. 259.
Jousse C, Pujos-Guillot E. Exploring metabolome with GC/MS. In: Rolin D, editor. Metabolomics coming of age with its technological diversity. 1st ed. London: Elsevier Science; 2012. p. 316.
Lamichhane S, Sen P, Dickens A, Hyötyläinen T, Orešič M. An overview of metabolomics data analysis: current tools and future perspectives. In: Jaumot J, Bedia C, Tauler R, editors. Data analysis for omic sciences: methods and applications. 1st ed. Amsterdam: Elsevier Science; 2018. p. 392.
Michael AF, Drummond KN, Doeden D, Anderson JA, Good RA. Tryptophan metabolism in man. J Clin Invest. 1964;43:1730–46.
Morita I, Kawamoto M, Hattori M, Eguchi K, Sekiba K, Yoshida H. Determination of tryptophan and its metabolites in human plasma and serum by high-performance liquid chromatography with automated sample clean-up system. J Chromatogr B Biomed Appl. 1990;526:367–74.
Young SN, Gauthier S. Effect of tryptophan administration on tryptophan, 5-hydroxyindoleacetic acid and indoleacetic acid in human lumbar and cisternal cerebrospinal fluid. J Neurol Neurosurg Psychiatry. 1981;44:323–8.
Brown DG, Rao S, Weir TL, O'Malia J, Bazan M, Brown RJ, et al. Metabolomics and metabolic pathway networks from human colorectal cancers, adjacent mucosa, and stool. Cancer Metab. 2016;4:11.
Goedert JJ, Sampson JN, Moore SC, Xiao Q, Xiong X, Hayes RB, et al. Fecal metabolomics: assay performance and association with colorectal cancer. Carcinogenesis. 2014;35:2089–96.
Ninomiya S, Tsurumi H, Hara T, Hoshi M, Goto N, Kanemura N, et al. Tryptophan catabolism is associated with clinical outcome of patients with malignant lymphoma. Blood. 2010;116:4146.
Nakamura N, Hara T, Shimizu M, Mabuchi R, Nagano J, Ohno T, et al. Effects of indoleamine 2,3-dioxygenase inhibitor in non-Hodgkin lymphoma model mice. Int J Hematol. 2015;102:327–34.
Litwack G. Chapter 8 - glycolysis and gluconeogenesis. In: Litwack G, editor. Human biochemistry. Boston: Academic Press; 2018. p. 183–98.
Bahado-Singh RO, Akolekar R, Mandal R, Dong E, Xia J, Kruger M, et al. Metabolomics and first-trimester prediction of early-onset preeclampsia. J Matern Fetal Neonatal Med. 2012;25:1840–7.
Scholl-Burgi S, Haberlandt E, Gotwald T, Albrecht U, Baumgartner Sigl S, Rauchenzauner M, et al. Stroke-like episodes in propionic acidemia caused by central focal metabolic decompensation. Neuropediatrics. 2009;40:76–81.
D’Aniello A, Vetere A, Fisher GH, Cusano G, Chavez M, Petrucelli L. Presence of d-alanine in proteins of normal and Alzheimer human brain. Brain Res. 1992;592:44–8.
Fukushima T, Santa T, Homma H, Nagatomo R, Imai K. Determination of D-amino acids in serum from patients with renal dysfunction. Biol Pharm Bull. 1995;18:1130–2.
Braverman ER, Pfeiffer CC, Blum K, Smayda R. The healing nutrients within: facts, findings, and new research on amino acids. 3rd ed. Laguna Beach: Basic Health Publications; 2003.
Chen KY, Liu X, Bu P, Lin CS, Rakhilin N, Locasale JW, et al. A metabolic signature of colon cancer initiating cells. Conf Proc IEEE Eng Med Biol Soc. 2014;2014:4759–62.
Strassman M, Ceci LN. Enzymatic formation of alpha-isopropylmalic acid, an intermediate in leucine biosynthesis. J Biol Chem. 1963;238:2445–52.
Ananieva EA, Wilkinson AC. Branched-chain amino acid metabolism in cancer. Curr Opin Clin Nutr Metab Care. 2018;21:64–70.
de Koning TJ, Snell K, Duran M, Berger R, Poll-The BT, Surtees R. L-serine in disease and development. Biochem J. 2003;371:653–61.
Hausen A, Wachter H. Pteridines in the assessment of neoplasia. J Clin Chem Clin Biochem. 1982;20:593–602.
Rokos H, Rokos K, Frisius H, Kirstaedter HJ. Altered urinary excretion of pteridines in neoplastic disease. Determination of biopterin, neopterin, xanthopterin, and pterin. Clin Chim Acta. 1980;105:275–86.
Dalla Pozza E, Dando I, Pacchiana R, Liboi E, Scupoli MT, Donadelli M, et al. Regulation of succinate dehydrogenase and role of succinate in cancer. Semin Cell Dev Biol. 2020;98:4–14.
Zhao T, Mu X, You Q. Succinate: an initiator in tumorigenesis and progression. Oncotarget. 2017;8:53819–28.
Battelli MG, Polito L, Bortolotti M, Bolognesi A. Xanthine oxidoreductase in cancer: more than a differentiation marker. Cancer Med. 2016;5:546–57.
Battelli MG, Bortolotti M, Polito L, Bolognesi A. Metabolic syndrome and cancer risk: the role of xanthine oxidoreductase. Redox Biol. 2019;21:101070.
Xu H, Li C, Mozziconacci O, Zhu R, Xu Y, Tang Y, et al. Xanthine oxidase-mediated oxidative stress promotes cancer cell-specific apoptosis. Free Radic Biol Med. 2019;139:70–9.
Yang HJ, Bogomolnaya L, McClelland M, Andrews-Polymenis H. De novo pyrimidine synthesis is necessary for intestinal colonization of Salmonella Typhimurium in chicks. PLoS One. 2017;12:e0183751.
Lee B, Singh RH, Rhead WJ, Sniderman King L, Smith W, Summar ML. Considerations in the difficult-to-manage urea cycle disorder patient. Crit Care Clin. 2005;21:S19–25.
Summar ML, Barr F, Dawling S, Smith W, Lee B, Singh RH, et al. Unmasked adult-onset urea cycle disorders in the critical care setting. Crit Care Clin. 2005;21:S1–8.
Laconi E, Denda A, Rao PM, Rajalakshmi S, Pani P, Sarma DS. Studies on liver tumor promotion in the rat by orotic acid: dose and minimum exposure time required for dietary orotic acid to promote hepatocarcinogenesis. Carcinogenesis. 1993;14:1771–5.
Ohshima M, Sugahara K, Kasahara K, Katakura A. Metabolomic analysis of the saliva of Japanese patients with oral squamous cell carcinoma. Oncol Rep. 2017;37:2727–34.
Nicolaides C, Fountzilas G, Zoumbos N, Skarlos D, Kosmidis P, Pectasides D, et al. Diffuse large cell lymphomas: identification of prognostic factors and validation of the international non-Hodgkin’s lymphoma prognostic index. A Hellenic Cooperative Oncology Group Study. Oncology. 1998;55:405–15.
Pellock SJ, Redinbo MR. Glucuronides in the gut: sugar-driven symbioses between microbe and host. J Biol Chem. 2017;292:8569–76.
Afratis N, Gialeli C, Nikitovic D, Tsegenidis T, Karousou E, Theocharis AD, et al. Glycosaminoglycans: key players in cancer cell biology and treatment. FEBS J. 2012;279:1177–97.
Bezabeh T, Ijare OB, Albiin N, Arnelo U, Lindberg B, Smith IC. Detection and quantification of D-glucuronic acid in human bile using 1H NMR spectroscopy: relevance to the diagnosis of pancreatic cancer. MAGMA. 2009;22:267–75.
Acknowledgments
Gustavo Henrique Bueno Duarte would like to thank National Council for Scientific and Technological Development (Project 870359/1997-5) for the scholarship, the São Paulo Research Foundation (Process 2016/07014-5) for the research financial support, and Agilent Technologies for technical support. Hans R. Zamora Obando would like to thank the University of Costa Rica for the scholarship (OAICE-159-2019).
Funding
This study was funded by the National Council for Scientific and Technological Development (grant number 870359/1997-5) and the São Paulo Research Foundation (grant number 2016/07014-5).
Author information
Authors and Affiliations
Contributions
Conceptualization: Gustavo Henrique Bueno Duarte, Ana Valéria Colnaghi Simionato, Marcos Nogueira Eberlin, Jayr Schmidt-Filho, Vladmir C. Cordeiro de Lima, Felipe D’Almeida Costa, Victor Piana Andrade; methodology: Gustavo Henrique Bueno Duarte, Ana Valéria Colnaghi Simionato; formal analysis and investigation: Gustavo Henrique Bueno Duarte; writing—original draft preparation: Gustavo Henrique Bueno Duarte; writing—review and editing: Gustavo Henrique Bueno Duarte, Anna Maria Alves de Piloto Fernandes, Álex Aparecido Rosini Silva, Hans Rolando Zamora Obando, Alan Gonçalves Amaral, Alessandra de Sousa Mesquita, Jayr Schmidt-Filho, Vladmir C. Cordeiro de Lima, Felipe D’Almeida Costa, Victor Piana Andrade, Andréia de Melo Porcari, Marcos Nogueira Eberlin, Ana Valéria Colnaghi Simionato; funding acquisition: Ana Valéria Colnaghi Simionato; resources: Ana Valéria Colnaghi Simionato; supervision: Ana Valéria Colnaghi Simionato, Marcos Nogueira Eberlin.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee (Research Ethics Committee of the Medical Sciences College at University of Campinas and AC Camargo Cancer Center CAEE 35481614.7.0000.5404), with the 1964 Helsinki declaration, and its later amendments or comparable ethical standards.
Consent to participate
Informed consent to participate was obtained from all individual participants included in the study.
Consent for publication
Informed consent for publication was obtained from all individual participants included in the study.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Bueno Duarte, G.H., de Piloto Fernandes, A.M.A., Silva, A.A.R. et al. Gas chromatography-mass spectrometry untargeted profiling of non-Hodgkin’s lymphoma urinary metabolite markers. Anal Bioanal Chem 412, 7469–7480 (2020). https://doi.org/10.1007/s00216-020-02881-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-020-02881-5