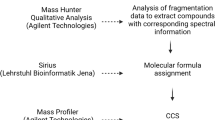
Abstract
Collision cross section (CCS) values are descriptors of the 3D structure of ions which can be determined by ion mobility spectrometry (IMS). Currently, most lipidomic studies involving CCS value determination concern eukaryote samples (e.g. human, bovine) and to a lower extent prokaryote samples (e.g. bacteria). Here, we report CCS values obtained from traveling wave ion mobility spectrometry (TWCCSN2) measurements from the bacterial membrane of Pseudomonas aeruginosa—a bacterium ranked as priority 1 for the R&D of new antibiotics by the World Health Organization. In order to cover the lack of reference compounds which could cover the m/z and CCS ranges of the membrane lipids of P. aeruginosa, three calibrants (polyalanine, dextran and phospholipids) were used for the TWCCSN2 calibration. A shift from the published lipid CCS values was systematically observed (ΔCCS% up to 9%); thus, we proposed a CCS correction strategy. This correction strategy allowed a reduction in the shift (ΔCCS%) between our measurements and published values to less than 2%. This correction was then applied to determine the CCS values of Pseudomonas aeruginosa lipids which have not been published yet. As a result, 32 TWCCSN2 values for [M+H]+ ions and 24 TWCCSN2 values for [M−H]− ions were obtained for four classes of phospholipids (phosphatidylethanolamines (PE), phosphatidylcholines (PC), phosphatidylglycerols (PG) and diphosphatidylglycerols—known as cardiolipins (CL)).

Graphical abstract



Similar content being viewed by others
References
Obritsch MD, Fish DN, MacLaren R, Jung R. Nosocomial infections due to multidrug-resistant Pseudomonas aeruginosa: epidemiology and treatment options. Pharmacotherapy. 2005;25(10):11.
Curran CS, Bolig T, Torabi-Parizi P. Mechanisms and targeted therapies for Pseudomonas aeruginosa lung infection. Am J Respir Crit Care Med. 2018;197(6):708–27. https://doi.org/10.1164/rccm.201705-1043SO.
Tacconelli E, Carrara E, Savoldi A, Harbarth S, Mendelson M, Monnet DL, et al. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis. 2018;18(3):318–27. https://doi.org/10.1016/s1473-3099(17)30753-3.
Bassetti M, Vena A, Croxatto A, Righi E, Guery B. How to manage Pseudomonas aeruginosa infections. Drugs Context. 2018;7:212527. https://doi.org/10.7573/dic.212527.
Escriba PV. Membrane-lipid therapy: a new approach in molecular medicine. Trends Mol Med. 2006;12(1):34–43. https://doi.org/10.1016/j.molmed.2005.11.004.
Gaviard C, Jouenne T, Hardouin J. Proteomics of Pseudomonas aeruginosa: the increasing role of post-translational modifications. Expert Rev Proteomics. 2018;15(9):757–72. https://doi.org/10.1080/14789450.2018.1516550.
Kamath KS, Krisp C, Chick J, Pascovici D, Gygi SP, Molloy MP. Pseudomonas aeruginosa proteome under hypoxic stress conditions mimicking the cystic fibrosis lung. J Proteome Res. 2017;16(10):3917–28. https://doi.org/10.1021/acs.jproteome.7b00561.
Hare NJ, Solis N, Harmer C, Marzook NB, Rose B, Harbour C, et al. Proteomic profiling of Pseudomonas aeruginosa AES-1R, PAO1 and PA14 reveals potential virulence determinants associated with a transmissible cystic fibrosis-associated strain. BMC Microbiol. 2012;12:16. https://doi.org/10.1186/1471-2180-12-16.
Benamara H, Rihouey C, Abbes I, Mlouka MAB, Hardouin J, Jouenne T, et al. Characterization of membrane lipidome changes in Pseudomonas aeruginosa during biofilm growth on glass wool. PloS One. 2014;9(9):9. https://doi.org/10.1371/journal.pone.0108478.g001.
Benamara H, Rihouey C, Jouenne T, Alexandre S. Impact of the biofilm mode of growth on the inner membrane phospholipid composition and lipid domains in Pseudomonas aeruginosa. Biochim Biophys Acta. 2011;1808(1):98–105. https://doi.org/10.1016/j.bbamem.2010.09.004.
Ewing MA, Glover MS, Clemmer DE. Hybrid ion mobility and mass spectrometry as a separation tool. J Chromatogr A. 2016;1439:3–25. https://doi.org/10.1016/j.chroma.2015.10.080.
Cumeras R, Figueras E, Davis CE, Baumbach JI, Gracia I. Review on ion mobility spectrometry. Part 1: current instrumentation. Analyst. 2015;140(5):1376–90. https://doi.org/10.1039/c4an01100g.
Gabelica V, Marklund E. Fundamentals of ion mobility spectrometry. Curr Opin Chem Biol. 2018;42:51–9. https://doi.org/10.1016/j.cbpa.2017.10.022.
Plante PL, Francovic-Fontaine E, May JC, McLean JA, Baker ES, Laviolette F, et al. Predicting ion mobility collision cross-sections using a deep neural network: DeepCCS. Anal Chem. 2019;91(8):5191–9. https://doi.org/10.1021/acs.analchem.8b05821.
Zhou Z, Tu J, Xiong X, Shen X, Zhu ZJ. LipidCCS: prediction of collision cross-section values for lipids with high precision to support ion mobility-mass spectrometry-based lipidomics. Anal Chem. 2017;89(17):9559–66. https://doi.org/10.1021/acs.analchem.7b02625.
Smith DP, Knapman TW, Campuzano I, Malham RW, Berryman JT, Radford SE, et al. Deciphering drift time measurements from travelling wave ion mobility spectrometry-mass spectrometry studies. Eur J Mass Spectrom (Chichester). 2009;15(2):113–30. https://doi.org/10.1255/ejms.947.
Paglia G, Astarita G. Metabolomics and lipidomics using traveling-wave ion mobility-mass spectrometry. Nat Protoc. 2017;12(4):17. https://doi.org/10.1038/nprot.2017.013.
Paglia G, Kliman M, Claude E, Geromanos S, Astarita G. Applications of ion-mobility mass spectrometry for lipid analysis. Anal Bioanal Chem. 2015;407:12.
Paglia G, Williams JP, Menikarachchi L, Thompson JW, Tyldesley-Worster R, Halldorsson S, et al. Ion mobility derived collision cross sections to support metabolomics applications. Anal Chem. 2014;86(8):3985–93. https://doi.org/10.1021/ac500405x.
Hines KM, May JC, McLean JA, Xu L. Evaluation of collision cross section calibrants for structural analysis of lipids by traveling wave ion mobility-mass spectrometry. Anal Chem. 2016;88(14):7329–36. https://doi.org/10.1021/acs.analchem.6b01728.
Levy AJ, Oranzi NR, Ahmadireskety A, Kemperman RHJ, Wei MS, Yost RA. Recent progress in metabolomics using ion mobility-mass spectrometry. TrAC Trends Anal Chem. 2019;116:274–81. https://doi.org/10.1016/j.trac.2019.05.001.
Tejada-Casado C, Hernandez-Mesa M, Monteau F, Lara FJ, Olmo-Iruela MD, Garcia-Campana AM, et al. Collision cross section (CCS) as a complementary parameter to characterize human and veterinary drugs. Anal Chim Acta. 2018;1043:52–63. https://doi.org/10.1016/j.aca.2018.09.065.
May JC, Morris CB, McLean JA. Ion mobility collision cross section compendium. Anal Chem. 2017;89(2):1032–44. https://doi.org/10.1021/acs.analchem.6b04905.
Schrimpe-Rutledge AC, Sherrod SD, McLean JA. Improving the discovery of secondary metabolite natural products using ion mobility-mass spectrometry. Curr Opin Chem Biol. 2018;42:160–6. https://doi.org/10.1016/j.cbpa.2017.12.004.
Hines KM, Xu L. Lipidomic consequences of phospholipid synthesis defects in Escherichia coli revealed by HILIC-ion mobility-mass spectrometry. Chem Phys Lipids. 2019;219:15–22. https://doi.org/10.1016/j.chemphyslip.2019.01.007.
Zheng X, Aly NA, Zhou Y, Dupuis KT, Bilbao A, Paurus VL, et al. A structural examination and collision cross section database for over 500 metabolites and xenobiotics using drift tube ion mobility spectrometry. Chem Sci. 2017;8(11):7724–36. https://doi.org/10.1039/c7sc03464d.
Ridenour WB, Kliman M, McLean JA, Caprioli RM. Structural characterization of phospholipids and peptides directly from tissue sections by MALDI traveling-wave ion mobility-mass spectrometry. Anal Chem. 2010;82(5):1881–9. https://doi.org/10.1021/ac9026115.
Jackson SN, Ugarov M, Post JD, Egan T, Langlais D, Schultz JA, et al. A study of phospholipids by ion mobility TOFMS. J Am Soc Mass Spectrom. 2008;19(11):1655–62. https://doi.org/10.1016/j.jasms.2008.07.005.
Harris RA, Leaptrot KL, May JC, McLean JA. New frontiers in lipidomics analyses using structurally selective ion mobility-mass spectrometry. TrAC Trends Anal Chem. 2019;116:316–23. https://doi.org/10.1016/j.trac.2019.03.031.
Hinz C, Liggi S, Griffin JL. The potential of ion mobility mass spectrometry for high-throughput and high-resolution lipidomics. Curr Opin Chem Biol. 2018;42:42–50. https://doi.org/10.1016/j.cbpa.2017.10.018.
Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37(8):7.
Hofmann J, Struwe WB, Scarff CA, Scrivens JH, Harvey DJ, Pagel K. Estimating collision cross sections of negatively charged N-glycans using traveling wave ion mobility-mass spectrometry. Anal Chem. 2014;86(21):10789–95. https://doi.org/10.1021/ac5028353.
Pagel K, Harvey DJ. Ion mobility-mass spectrometry of complex carbohydrates: collision cross sections of sodiated N-linked glycans. Anal Chem. 2013;85(10):5138–45. https://doi.org/10.1021/ac400403d.
Bush MF, Campuzano ID, Robinson CV. Ion mobility mass spectrometry of peptide ions: effects of drift gas and calibration strategies. Anal Chem. 2012;84(16):7124–30. https://doi.org/10.1021/ac3014498.
Domalain V, Hubert-Roux M, Tognetti V, Joubert L, Lange CM, Rouden J, et al. Enantiomeric differentiation of aromatic amino acids using traveling wave ion mobility-mass spectrometry. Chem Sci. 2014;5(8):3234–9. https://doi.org/10.1039/c4sc00443d.
Garrett TA, Kordestani R, Raetz CRH. Quantification of cardiolipin by liquid chromatography-electrospray ionization mass spectrometry. Method. Enzymol. 2007;433:213–30. https://doi.org/10.1016/s0076-6879(07)33012-7.
Liebisch G, Vizcaino JA, Kofeler H, Trotzmuller M, Griffiths WJ, Schmitz G, et al. Shorthand notation for lipid structures derived from mass spectrometry. J Lipid Res. 2013;54(6):1523–30. https://doi.org/10.1194/jlr.M033506.
Groessl M, Graf S, Knochenmuss R. High resolution ion mobility-mass spectrometry for separation and identification of isomeric lipids. Analyst. 2015;140(20):6904–11. https://doi.org/10.1039/c5an00838g.
Leaptrot KL, May JC, Dodds JN, McLean JA. Ion mobility conformational lipid atlas for high confidence lipidomics. Nat Commun. 2019;10(1):985. https://doi.org/10.1038/s41467-019-08897-5.
Chao J, Wolfaardt GM, Arts MT. Characterization of Pseudomonas aeruginosa fatty acid profiles in biofilms and batch planktonic cultures. Can J Microbiol. 2010;56(12):1028–39. https://doi.org/10.1139/W10-093.
Schlame M. Cardiolipin synthesis for the assembly of bacterial and mitochondrial membranes. J Lipid Res. 2008;49(8):1607–20. https://doi.org/10.1194/jlr.R700018-JLR200.
Kim HI, Kim H, Pang ES, Ryu EK, Beegle LW, Loo JA, et al. Structural characterization of unsaturated phosphatidylcholines using traveling wave ion mobility spectrometry. Anal Chem. 2009;81(20):8289–97. https://doi.org/10.1021/ac900672a.
El Khoury M, Swain J, Sautrey G, Zimmermann L, Van Der Smissen P, Décout J-L, et al. Targeting bacterial cardiolipin enriched microdomains: an antimicrobial strategy used by amphiphilic aminoglycoside antibiotics. Sci Rep. 2017;7(1). https://doi.org/10.1038/s41598-017-10543-3.
Funding
The authors gratefully acknowledge European Regional Development Fund (ERDF, no. HN0001343), Labex SynOrg (ANR-11-LABX-0029) and Région Normandie for their financial support.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Deschamps, E., Schmitz-Afonso, I., Schaumann, A. et al. Determination of the collision cross sections of cardiolipins and phospholipids from Pseudomonas aeruginosa by traveling wave ion mobility spectrometry-mass spectrometry using a novel correction strategy. Anal Bioanal Chem 411, 8123–8131 (2019). https://doi.org/10.1007/s00216-019-02194-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-019-02194-2