Abstract
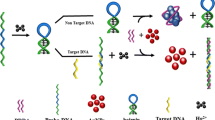
A novel electrochemical luminescence (ECL) aptamer biosensor via polymerase amplification is constructed for label-free detection of leukemia marker mRNA (miR-16). In order to achieve the ultrasensitive detection of the target mRNA, the cyclic target chain displacement polymerization of leukemia marker mRNA assisted with Klenow fragment of DNA polymerase is employed. The determination is carried out by recording the ECL emission of pyridine ruthenium (Ru(bpy)32+) complexes embedded into the assistance DNA (ADNA) loaded on the nanogold surface, after the hybridization reaction between the probe DNA (PDNA) and the remaining sequence of the CP’s stem part, and the formation of a core-shell sun-like structure. The mercapto-modified capture DNA (CP) is immobilized on the surface of a magneto-controlled glassy carbon electrode by Au-S bond. The CP is opened and hybridized with the target mRNA to form double-stranded DNA. In the presence of polymerase, primer DNA, and bases (dNTPs), the primer chain gets access to its complementary sequence of the stem part and then triggers a polymerization of the DNA strand, leading to the release of mRNA and starting the next polymerization cycle. Finally, the composite of PDNA-covered and ADNA-covered (embedded with Ru(bpy)32+) gold nanoparticles (hereafter called AuNPs@(PDNA+ADNA-Ru(bpy)32+) is added, and the ECL intensity is recorded. Because of the polymerization cycle and the aggregation of the illuminator of Ru(bpy)32+, the detected signal is amplified significantly. The results showed that the corresponding ECL signal has a good linear relationship with a logarithm of target mRNA concentration in the range of 1 × 10−16 to 1 × 10−7 mol/L, with a detection limit of 4.3 × 10−17 mol/L. The mRNA spiked in the human serum sample is determined, and the recoveries are from 97.2 to 102.0%. This sensor demonstrates good selectivity, stability, and reproducibility.

ᅟ








Similar content being viewed by others
References
Musto P, Negrini M, De LL, Cuneo A. Dissecting chronic lymphocytic leukemia with 13q-using microRNA expression profile: editorial for the paper “miRNA expression profile of chronic lymphocytic leukemia patients with 13q deletion”. Leuk Res. 2016;47:114–5.
Achille NJ, Othus M, Phelam K, Zhang SB, Cooper K, Godwin JE, et al. Association between early promoter-specific DNA methylation changes and outcome in older acute myeloid leukemia patients. Leuk Res. 2016;42:68–74.
Buckley SA, Jimenez SD, Othus M, Walter RB, Lee SJ. Quality of life from the perspective of the patient with acute myeloid leukemia. Cancer. 2018;124:145–52.
Setiadi A, Owen D, Tsang A, Milner R, Vercautern S. The significance of peripheral blood minimal residual disease to predict early disease response in patients with B–cell acute lymphoblastic leukemia. Int J Lab Hematol. 2016;38:527–34.
Boyerinas B, Zafrir M, Yesilkanal AE, Price TT, Hyjek EM, Sipkins DA. Adhesion to osteopontin in the bone marrow niche regulates lymphoblastic leukemia cell dormancy. Blood. 2013;121:4821–31.
Jenderny J, Goldmann C, Thede R, Ebrecht M, Korioth F. Detection of clonal aberrations by cytogenetic analysis after different culture methods and by FISH in 129 patients with chronic lymphocytic leukemia. Cytogenet Genome Res. 2014;144:163–8.
Esfandyarpour R, Yang L, Koochak Z, Harris JS, Davis RW. Nanoelectronic three-dimensional (3D) nanotip sensing array for real-time, sensitive, label-free sequence specific detection of nucleic acids. Biomed Microdevices. 2016;18:1–10.
Kitamura M, Aragane M, Nakamura K, Watanabe K, Sasaki Y. Development of loop-mediated isothermal amplification (LAMP) assay for rapid detection of Cannabis sativa. Biol Pharm Bull. 2016;39:1144–9.
Zhang M, Hai H, Zhou FY, Zhong JC, Li JP. Electrochemical luminescent DNA sensor based on polymerase-assisted signal amplification. Chin J Anal Chem. 2018;46:203–9.
Martens ES, Böttcher R, Croce CM, Jenster G, Visakorpi T, Calin GA. Long noncoding RNA in prostate, bladder, and kidney cancer. Eur Urol. 2014;65:1140–51.
Yang LM, Li J, Pan W, Wang HY, Li N, Tang BY. Fluorescence and photoacoustic dual-mode imaging of tumor-related mRNA with a covalent linkage-based DNA nanoprobe. Chem Commun. 2018;54:3656–9.
Miao P, Jiang YT, Zhang T, Huang Y, Yang YG. Electrochemical sensing of attomolar miRNA combining cascade strand displacement polymerization and reductant-mediated amplification. Chem Commun. 2018;54:7366–9.
Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci. 2002;99:15524–9.
Enfield KSS, Martinez VD, Marshall EA, Stewart GL, Kung SH, Enterina JR, et al. Deregulation of small non-coding RNAs at the DLK1-DIO3 imprinted locus predicts lung cancer patient outcome. Oncotarget. 2016;7:80957–66.
Yu MM, Yao S, Luo KM, Mu QF, Yu Y, Luo GH, et al. Apolipoprotein M increases the expression of vitamin D receptor mRNA in colorectal cancer cells detected with duplex fluorescence reverse transcription-quantitative polymerase chain reaction. Mol Med Rep. 2017;16:1167–72.
Sanyal S, Siriwardena AK, Byers R. Measurement of indicator genes using global complementary DNA (cDNA) amplification, by polyadenylic acid reverse transcriptase polymerase chain reaction (poly A RT-PCR): a feasibility study using paired samples from tissue and ductal juice in patients undergoing pancreatoduodenectomy. Pancreatology. 2018;18:458–62.
Lin X, Chen Y. Identification of potentially functional circRNA-miRNA-mRNA regulatory network in hepatocellular carcinoma by integrated microarray analysis. Med Sci Monit. 2018;24:70–8.
Ma Y, Shi L, Zheng C. Microarray analysis of lncRNA and mRNA expression profiles in mice with allergic rhinitis. Int J Pediatr Otorhinolaryngol. 2018;104:58–65.
Wang S, Zhang LQ, Wan S, Gansiz S, Cui C, Liu Y, et al. Aptasensor with expanded nucleotide using DNA nanotetrahedra for electrochemical detection of cancerous exosomes. ACS Nano. 2017;11:3943–9.
Li JP, Sun M, Wei XP, Zhang LM, Zhang Y. An electrochemical aptamer biosensor based on “gate-controlled” effect using β-cyclodextrin for ultra-sensitive detection of trace mercury. Biosens Bioelectron. 2015;74:423–6.
Zhou FY, Hai H, Yuan YL, Li JP. Ultrasensitive electrochemiluminescence biosensor for mRNA based on polymerase assisted signal amplification. Electroanalysis. 2017;29:983–9.
Qin GX, Zhao SL, Huang Y, Jiang J, Liu YM. A sensitive gold nanoparticles sensing platform based on resonance energy transfer for chemiluminescence light on detection of biomolecules. Biosens Bioelectron. 2013;46:119–23.
Wang LP, Huang YB, Lai YH. Surface enhanced Raman scattering activity of dual-functional Fe3O4/Au composites. Appl Surf Sci. 2018;435:290–6.
Yao Y, Xue M, Zhang ZB, Zhang MM, Wang Y, Huang FH. Gold nanoparticles stabilized by an amphiphilic pillar [5] arene: preparation, self-assembly into composite microtubes in water and application in green catalysis. Chem Sci. 2013;4:3667–72.
Ding QW, Zhan QQ, Zhou XM, Zhang T, Xing D. Theranostic upconversion nanobeacons for tumor mRNA ratiometric fluorescence detection and imaging-monitored drug delivery. Small. 2016;12:5944–53.
Ali MM, Li F, Zhang ZQ, Zhang KX, Kang DK, Ankrum JA, et al. Rolling circle amplification: a versatile tool for chemical biology, materials science and medicine. Chem Soc Rev. 2014;43:3324–41.
Alsage OA, Kumar S, Zhu BC, Sejdic JT, McNatty KP, Hodgkiss JM. Ultrasensitive colorimetric detection of 17β-estradiol: the effect of shortening DNA aptamer sequences. Anal Chem. 2015;87:4201–9.
Zhao FL, Xie QY, Xu MF, Wang SY, Zhou JY, Liu F. RNA aptamer based electrochemical biosensor for sensitive and selective detection of cAMP. Biosens Bioelectron. 2015;66:238–43.
Funding
This research is financially supported by the National Natural Science Foundation of China (No.21765006), the Guangxi Natural Science Foundation, China (No. 2015GXNSFFA139005), and the High-Level Innovation Teams of Guangxi Colleges and Universities and Outstanding Scholars Program (Guijiaoren[2014]49).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical standards and informed consent
The study and experimental sections were approved by the Hospital of Guilin University of Technology. Human serum samples used in this study do not have any identifying information about all the participants that provided written informed consent.
Electronic supplementary material
ESM 1
(PDF 204 kb)
Rights and permissions
About this article
Cite this article
Zhang, M., Zhou, F., Zhou, D. et al. An aptamer biosensor for leukemia marker mRNA detection based on polymerase-assisted signal amplification and aggregation of illuminator. Anal Bioanal Chem 411, 139–146 (2019). https://doi.org/10.1007/s00216-018-1424-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-018-1424-9