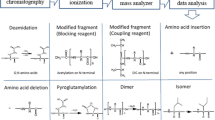
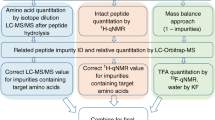
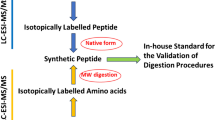
Abstract
Identification and quantitation of related impurities is vital in obtaining corrected purity values for peptide certified reference materials. The sensitivity and selectivity of high-resolution mass spectrometry (MS) renders it an indispensable technique in this arena. Typical quantitation efforts involve constructing external calibration curves, although analysis of dilute peptide solutions can be complicated by analyte adsorption to vial walls, instrument tubing, etc. The standard addition method alleviates many concerns associated with this sample loss as the calibrant solutions more closely match the matrix of the samples. Yet, both strategies require acquisition of synthetic impurity peptide standards. Label-free proteomics relies on electrospray ionization (ESI)-MS signals to quantify identical peptides across multiple samples; however, peptides of differing sequence can exhibit widely disparate ESI-MS responses. This study explores the use of peak area ratios to quantitate sequence-related peptide impurities in an angiotensin II candidate certified reference material. Using synthetic standards of five abundant substances, impurity mass fractions calculated via the relative response method are in reasonable agreement with those determined from standard addition experiments, whereas external calibration measurements frequently overestimate impurity amounts. For a synthetic peptide and its related sequence impurities, the relative response method can expedite analysis and lower expenditures, and in some cases improve data quality.





Similar content being viewed by others
References
Zeng K, Geerlof-Vidavisky I, Gucinsky A, Jiang X, Boyne MT II. Liquid chromatography-high resolution mass spectrometry for peptide drug quality control. AAPS J. 2015;17:643–51. https://doi.org/10.1208/s12248-015-9730-z.
Percy AJ, Yang J, Hardie DB, Chambers AG, Tamura-Wells J, Borchers CH. Precise quantitation of 136 urinary proteins by LC/MRM-MS using stable isotope labeled peptides as internal standards for biomarker discovery and/or verification studies. Methods. 2015;81:24–33. https://doi.org/10.1016/j.ymeth.2015.04.001.
Goebel-Stengel M, Stengel A, Taché Y, Reeve JR Jr. The importance of using optimal plasticware and glassware in studies involving peptides. Anal Biochem. 2011;414:38–46. https://doi.org/10.1016/j.ab.2011.02.009.
Stejskal K, Potěšil D, Zdráhal Z. Suppression of peptide losses in autosampler vials. J Proteome Res. 2013;12:3057–62. https://doi.org/10.1021/pr400183v.
Megger DA, Bracht T, Meyer HE, Sitek B. Label-free quantification in clinical proteomics. Biochim Biophys Acta, Proteins Proteomics. 2013;1834:1581–90. https://doi.org/10.1016/j.bbapap.2013.04.001.
Nahnsen S, Bielow C, Reinert K, Kohlbacher O. Tools for label-free peptide quantification. Mol Cell Proteomics. 2013;12:549–56. https://doi.org/10.1074/mcp.R112.025163.
Navarro P, Kuharev J, Gillet LC, Bernhardt OM, MacLean B, Rost HL, et al. A multicenter study benchmarks software tools for label-free proteome quantification. Nat Biotechnol. 2016;34:1130–6. https://doi.org/10.1038/nbt.3685.
Cech NB, Enke CG. Practical implications of some recent studies in electrospray ionization fundamentals. Mass Spectrom Rev. 2001;20:362–87. https://doi.org/10.1002/mas.10008.
Melanson JE, Thibeault M-P, Stocks BB, Leek DM, McRae G, Meija J. Purity assignment for peptide certified reference materials by combining the qNMR and LC-MS/MS amino acid analysis results: application to angiotensin II. Anal Bioanal Chem. https://doi.org/10.1007/s00216-018-1272-7.
Kościelniak P, Kozak J. Review of univariate standard addition calibration procedures in flow analysis. Crit Rev Anal Chem. 2006;36:27–40. https://doi.org/10.1080/10408340500451999.
Chalkley RJ, Baker PR, Medzihradszky KF, Lynn AJ, Burlingame AL. In-depth analysis of tandem mass spectrometry data from disparate instrument types. Mol Cell Proteomics. 2008;7:2386–98. https://doi.org/10.1074/mcp.M800021-MCP200.
Meija J, Pagliano E, Mester Z. Coordinate swapping in standard addition graphs for analytical chemistry: a simplified path for uncertainty calculation in linear and nonlinear plots. Anal Chem. 2014;86:8563–7. https://doi.org/10.1021/ac5014749.
Vahidi S, Konermann L. Strategies for minimizing spurious in-source CID for peptides during ESI-MS. In: 62nd ASMS Conference on Mass Spectrometry and Allied Topics, Baltimore, MD, 2014.
Liu C, Topchiy E, Lehmann T, Basile F. Characterization of the dehydration products due to thermal decomposition of peptides by liquid chromatography-tandem mass spectrometry. J Mass Spectrom. 2015;50:625–32. https://doi.org/10.1002/jms.3570.
Chelius D, Jing K, Lueras A, Rehder DS, Dillon TM, Vizel A, et al. Formation of pyroglutamic acid from N-terminal glutamic acid in immunoglobulin gamma antibodies. Anal Chem. 2006;78:2370–6. https://doi.org/10.1021/ac051827k.
Liu YD, Goetze AM, Bass RB, Flynn GC. N-terminal glutamate to pyroglutamate conversion in vivo for human IgG2 antibodies. J Biol Chem. 2011;286:11211–7. https://doi.org/10.1074/jbc.M110.185041.
Harris RJ, Kabakoff B, Macchi FD, Shen FJ, Kwong M, Andya JD, et al. Identification of multiple sources of charge heterogeneity in a recombinant antibody. J Chromatogr B. 2001;752:233–45. https://doi.org/10.1016/S0378-4347(00)00548-X.
Kumar M, Chatterjee A, Khedkar AP, Kusumanchi M, Adhikary L. Mass spectrometric distinction of in-source and in-solution pyroglutamate and succinimide in proteins: a case study on rhG-CSF. J Am Soc Mass Spectrom. 2012;24:202–12. https://doi.org/10.1007/s13361-012-0531-7.
Xie B, Sharp JS. Relative quantification of sites of peptide and protein modification using size exclusion chromatography coupled with electron transfer dissociation. J Am Soc Mass Spectrom. 2016;27:1322–7. https://doi.org/10.1007/s13361-016-1403-3.
Yang Y. Side reactions in peptide synthesis. London: Elsevier; 2016. https://doi.org/10.1016/B978-0-12-801009-9.00009-4.
Jia C, Lietz CB, Yu Q, Li L. Site-specific characterization of D-amino acid containing peptide epimers by ion mobility spectrometry. Anal Chem. 2014;86:2972–81. https://doi.org/10.1021/ac4033824.
Meija J, Coplen TB, Berglund M, Brand WA, De Bièvre P, Gröning M, et al. Isotopic compositions of the elements 2013 (IUPAC technical report). Pure Appl Chem. 2016;88:293–306. https://doi.org/10.1515/pac-2015-0503.
Rappold BA, Hoofnagle AN. Bias due to isotopic incorporation in both relative and absolute protein quantitation with carbon-13 and nitrogen-15 labeled peptides. Clin Mass Spectrom. 2017;3:13–21. https://doi.org/10.1016/j.clinms.2017.04.002.
Midani FS, Wynn ML, Schnell S. The importance of accurately correcting for the natural abundance of stable isotopes. Anal Biochem. 2017;520:27–43. https://doi.org/10.1016/j.ab.2016.12.011.
Enke CG. A predictive model for matrix and analyte effects in electrospray ionization of singly-charged ionic analytes. Anal Chem. 1997;69:4885–93. https://doi.org/10.1021/ac970095w.
Ahadi E, Konermann L. Modeling the behavior of coarse-grained polymer chains in charged water droplets: implication for the mechanism of electrospray ionization. J Phys Chem B. 2012;116:104–12. https://doi.org/10.1021/jp209344z.
Cech NB, Enke CG. Relating electrospray ionization response to nonpolar character of small peptides. Anal Chem. 2000;72:2717–23. https://doi.org/10.1021/ac9914869.
Gao Y, Wang Y. A method to determine the ionization efficiency change in peptides caused by phosphorylation. J Am Soc Mass Spectrom. 2007;18:1973–6. https://doi.org/10.1016/j.jasms.2007.08.010.
Stoppacher N, Josephs RD, Daireaux A, Choteau T, Westwood S, Wielgosz RI. Accurate quantification of impurities in pure peptide material—angiotensin I: comparison of calibration requirements and method performance characteristics of liquid chromatography coupled to hybrid tandem mass spectrometry and linear ion trap high-resolution mass spectrometry. Rapid Commun Mass Spectrom. 2015;29:1651–60. https://doi.org/10.1002/rcm.7261.
Pritchard C, O’Connor G, Ashcroft AE. The role of ion mobility spectrometry-mass spectrometry in the analysis of protein reference standards. Anal Chem. 2013;85:7205–12. https://doi.org/10.1021/ac400927s.
van Midwoud PM, Rieux L, Bischoff R, Verpoorte E, Niederländer HAG. Improvement of recovery and repeatability in liquid chromatography-mass spectrometry analysis of peptides. J Proteome Res. 2007;6:781–91. https://doi.org/10.1021/pr0604099.
Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, et al. Protein identification and analysis tools on the ExPASy server. In: Walker JM, editor. The proteomics protocols handbook. Totowa: Humana; 2005. p. 571–607. https://doi.org/10.1385/1-59259-890-0:571.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Stocks, B.B., Thibeault, MP., Meija, J. et al. Assessing MS-based quantitation strategies for low-level impurities in peptide reference materials: application to angiotensin II. Anal Bioanal Chem 410, 6963–6972 (2018). https://doi.org/10.1007/s00216-018-1302-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-018-1302-5