Abstract
In this work, a new approach is proposed to verify the differentiating characteristics of five bacteria (Escherichia coli, Enterococcus faecalis, Streptococcus salivarius, Streptococcus oralis, and Staphylococcus aureus) by using digital images obtained with a simple webcam and variable selection by the Successive Projections Algorithm associated with Linear Discriminant Analysis (SPA-LDA). In this sense, color histograms in the red–green–blue (RGB), hue-saturation-value (HSV), and grayscale channels and their combinations were used as input data, and statistically evaluated by using different multivariate classifiers (Soft Independent Modeling by Class Analogy (SIMCA), Principal Component Analysis-Linear Discriminant Analysis (PCA-LDA), Partial Least Squares Discriminant Analysis (PLS-DA) and Successive Projections Algorithm-Linear Discriminant Analysis (SPA-LDA)). The bacteria strains were cultivated in a nutritive blood agar base layer for 24 h by following the Brazilian Pharmacopoeia, maintaining the status of cell growth and the nature of nutrient solutions under the same conditions. The best result in classification was obtained by using RGB and SPA-LDA, which reached 94 and 100 % of classification accuracy in the training and test sets, respectively. This result is extremely positive from the viewpoint of routine clinical analyses, because it avoids bacterial identification based on phenotypic identification of the causative organism using Gram staining, culture, and biochemical proofs. Therefore, the proposed method presents inherent advantages, promoting a simpler, faster, and low-cost alternative for bacterial identification.

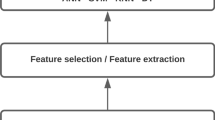
Summary of the new proposed methodology for bacteria classification by using color histograms and SPA-LDA





Similar content being viewed by others
References
van der Merwe RG, van Helden PD, Warren RM, Sampson SL, van Pittius NCG (2014) Phage-based detection of bacterial pathogens. Analyst. doi:10.1039/C4AN00208C
Hossain Z (2014) In: Motarjemi Y (ed) Encyclopedia of food safety, 1st edn. Academic, San Diego
Codina MG, de Cueto M, Vicente D, Echevarría JE, Prats G (2011) Microbiological diagnosis of central nervous system infections. Enferm Infecc Microbiol Clin 29:127–134
Guibet F, Amiel C, Cadot P, Cordevant C, Desmonts MH, Lange M, Marecat A, Travert J, Denis C, Mariey L (2003) Discrimination and classification of Enterococci by Fourier transform infrared (FT-IR) spectroscopy. Vib Spectrosc 33:133–142
Schleifer KH (2009) Classification of bacteria and Archaea: past, present and future. Syst Appl Microbiol 32:533–542
Xiao D, Zhao F, Lv M, Zhang H, Zhang Y, Huang H, Su P, Zhang Z, Zhang J (2012) Rapid identification of microorganisms isolated from throat swab specimens of community-acquired pneumonia patients by two MALDI-TOF MS systems. Diagn Microbiol Infect Dis 73:301–307
Nakai S, Wang ZH, Dou J, Nakamura S, Ogawa M, Nakai E, Vanderstoep J (1999) Gas chromatography/principal component similarity system for detection of E. coli and S. aureus contaminating salmon and hamburger. J Agric Food Chem 47:576–583
Li D, Truong TV, Bills TM, Holt BC, Van Derwerken DN, Williams JR, Acharya A, Robison RA, Tolley HD, Lee ML (2012) GC/MS method for positive detection of Bacillus anthracis endospores. Anal Chem 84:1637–1644
Hantula J, Kurki A, Vuoriranta P, Bamford DH (1991) Rapid classification of bacterial strains by SDS-polyacrylamide gel electrophoresis: population dynamics of the dominant dispersed phase bacteria of activated sludge. Appl Microbiol Biotechnol 34:551–555
Veloo ACM, Erhard M, Welker M, Welling GW, Degener JE (2011) Identification of Gram-positive anaerobic cocci by MALDI-TOF mass spectrometry. Syst Appl Microbiol 34:58–62
Beier BD, Quivey RG Jr, Berger AJ (2010) Identification of different bacterial species in biofilms using confocal Raman microscopy. J Biomed Opt 15:066001
Oust A, Møretrø T, Kirschner C, Narvhus JA, Kohler A (2004) FT-IR spectroscopy for identification of closely related lactobacilli. J Microbiol Methods 59:149–162
Preisner O, Lopes JA, Menezes JC (2008) Uncertainty assessment in FT-IR spectroscopy based bacteria classification models. Chemom Intell Lab Syst 94:33–42
Marques AS, de Melo MCN, Cidral TA, de Lima KMG (2014) Feature selection strategies for identification of Staphylococcus aureus recovered in blood cultures using FT-IR spectroscopy successive projections algorithm for variable selection: a case study. J Microbiol Methods 98:26–30
Giana HE, Silveira L Jr, Zângaro RA, Pacheco MTT (2003) Rapid identification of bacterial species by fluorescence spectroscopy and classification through principal components analysis. J Fluoresc 13:489–493
Sohn M, Himmelsbach DS, Barton FE, Fedorka-Cray PJ (2009) Fluorescence spectroscopy for rapid detection and classification of bacterial pathogens. Appl Spectrosc 63:1251–1255
Kim SW, Ban SH, Ahn CY, Oh HM, Chung H, Cho SH, Park YM, Liu JR (2006) Taxonomic discrimination of cyanobacteria by metabolic fingerprinting using proton nuclear magnetic resonance spectra and multivariate statistical analysis. J Plant Biol 49:271–275
Dubuisson M-P, Jain AK, Jain MK (1994) Segmentation and classification of bacterial culture images. J Microbiol Methods 19:279–295
Kumar S, Mittal GS (2008) Geometric and optical characteristics of five microorganisms for rapid detection using image processing. Biosyst Eng 99:1–8
Huff K, Aroonnual A, Littlejohn AE, Rajwa B, Bae E, Banada PP, Patsekin V, Hirleman ED, Robinson JP, Richards GP, Bhunia AK (2012) Light-scattering sensor for real-time identification of Vibrio parahaemolyticus, Vibrio vulnificus and Vibrio cholerae colonies on solid agar plate. Microb Biotechnol 5:607–620
Suchwałko A, Buzalewicz I, Wieliczko A, Podbielska H (2013) Bacteria species identification by the statistical analysis of bacterial colonies Fresnel patterns. Opt Express 21:11322–11337
Diniz PHGD, Dantas HV, Melo KDT, Barbosa MF, Harding DP, Nascimento ECL, Pistonesi MF, Band BSF, Araújo MCU (2012) Using a simple digital camera and SPA-LDA modeling to screen teas. Anal Methods 4:2648–2652
Domínguez MA, Diniz PHGD, Di Nezio MS, Araújo MCU, Centurión ME (2014) Geographical origin classification of Argentinean honeys using a digital image-based flow-batch system. Microchem J 112:104–108
Milanez KDTM, Pontes MJC (2014) Classification of edible vegetable oil using digital image and pattern recognition techniques. Microchem J 113:10–16
Soares SFC, Gomes AA, Galvão Filho AR, Araújo MCU, Galvão RKH (2013) The successive projections algorithm. Trends Anal Chem 42:84–98
Agência Nacional de Vigilância Sanitária (2010) Farmacopeia brasileira, Brasil
Clinical and Laboratory Standards Institute (2005) CLSI document M100-S15, EUA
Kennard RW, Stone LA (1969) Computer aided design of experiments. Technometrics 11:137–148
Ballabio D, Consonni V (2013) Classification tools in chemistry. Part 1: linear models. PLS-DA. Anal Methods 5:3790–3798
Acknowledgments
The authors gratefully acknowledge the Universidade Estadual da Paraíba for the financial support. The authors thank also Capes and CNPq Brazil scholarships and research fellowships. Paulo Henrique Gonçalves Dias Diniz also thanks to Fundação de Apoio à Pesquisa do Estado da Paraíba – FAPESQ-PB.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
de Almeida, V.E., da Costa, G.B., de Sousa Fernandes, D.D. et al. Using color histograms and SPA-LDA to classify bacteria. Anal Bioanal Chem 406, 5989–5995 (2014). https://doi.org/10.1007/s00216-014-8015-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-014-8015-1