Abstract
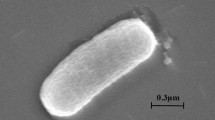
A gram-stain-negative, non-motile and rod-shaped strain, designated wg1T, was isolated from activated sludge obtained from wastewater treatment plant in Binzhou (Shandong province, PR China). Growth of strain wg1T occurred at 25–45 °C (optimum, 37 °C), at pH 7.0–9.0 (optimum growth at pH 8.0) and at a salinity range of 0–4% (optimum, 1%). The chemotaxonomic, phenotypic and genomic traits were investigated. The 16S rRNA gene sequence analysis showed that strain wg1T belonged to the genus Paracoccus. The species with highest similarity to strain wg1T was Paracoccus communis VKM B-2787T (98.27%), followed by Paracoccus kondratievae VKM B-2222T (98.25%). The isoprenoid quinone was Q-10. Major cellular fatty acids were summed feature 8, C16:0 and C18:0. The major polar lipids were diphosphatidylglycerol (DPG), phosphatidylethanolamine (PE), aminoglycolipid (AGL), phosphatidylglycerol (PG), phosphatidylcholine (PC), aminolipid (AL), one unidentified lipid (L) and one unidentified phospholipid (PL). The genome size was 4,834,448 bp with a G+C content of 67.67 mol%. The prediction result of secondary metabolites based on genome has shown that the strain wg1T contained 12 clusters, and the gene involved in primary metabolism showed differences in the comparison between wg1T and reference strains. The dDDH values of strain wg1T with P. communis VKM B-2787T, P. kondratievae VKM B-2222T and P. denitrificans DSM 413T were 45.30, 30.60 and 39.50%, respectively. Based on its physiological properties, chemotaxonomic characteristics and low ANI and dDDH results, strain wg1T is considered to represent a novel species for which the name Paracoccus binzhouensis sp. nov., is proposed. The type strain is wg1T (= KCTC 72861T = CCTCC AB 2019400T).

Similar content being viewed by others
Abbreviations
- PO:
-
Propylene oxide
- ANI:
-
Average nucleotide identity
- dDDH:
-
Digital DNA–DNA hybridization
- HPLC:
-
High performance liquid chromatography
- MIDI:
-
Microbial identification system
- KCTC:
-
Korean collection for type cultures
- CCTCC:
-
China Center for Type Culture Collection
- TLC:
-
Thin layer chromatography
References
Blin K, Shaw S, Steinke K, Villebro R, Ziemert N, Lee SY, Medema MH, Weber T (2019) antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res 47:W81–W87
Bondoc JMG, Wolf NM, Ndichuck M, Abad-Zapatero C, Movahedzadeh F (2017) Mutagenesis of threonine to serine in the active site of Mycobacterium tuberculosis fructose-1,6-bisphosphatase (Class II) retains partial enzyme activity. Biotechnol Rep 15:48–54
Bowman JP (2000) Description of Cellulophaga algicola sp. nov., isolated from the surfaces of Antarctic algae, and reclassification of Cytophaga uliginosa (ZoBell and Upham 1944) Reichenbach 1989 as Cellulophaga uliginosa comb. Int J Syst Evol Microbiol 50:1861–1868
Chun J, Oren A, Ventosa A, Christensen H, Arahal DR, Costa MS, Rooney AP, Xu XW, Meyer SD, Trujillo ME (2018) Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int J Syst Evol Microbiol 68:461–466
Collins MD, Goodfellow M, Minnikin DE (1980) Fatty acid, isoprenoid quinone and polar lipid composition in the classification of Curtobacterium and related taxa. J Gen Microbiol 118:29–37
Cui XL, Mao PH, Zeng M, Li WJ, Zhang LP, Xu LH, Jiang CL (2001) Streptimonospora salina gen. nov., sp. nov., a new member of the family Nocardiopsaceae. Int J Syst Evol Microbiol 51:357–363
Davis DH, Doudoroff M, Stanier RY, Mandel M (1969) Proposal to reject the genus Hydrogenomonas: taxonomic implications. Int J Syst Bacteriol 19:375–390
Dong X, Zhang G, Xiong Q, Liu D, Wang D, Liu Y, Wu G, Li P, Luo Y, Zhang R (2018) Paracoccus salipaludis sp. nov., isolated from saline–alkaline soil. Int J Syst Evol Microbiol 68:3812–3817
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Lee M, Woo SG, Park G, Kim MK (2011) Paracoccus caeni sp. nov., isolated from sludge. Int J Syst Evol Microbiol 61:1968–1972
Liu YL, Meng D, Li RR, Gu PF, Fan XY, Huang ZS, Du ZJ, Li Q (2019) Rhodoligotrophos defluvii sp. nov., isolated from activated sludge. Int J Syst Evol Microbiol 69:3830–3836
Liu ZP, Wang BJ, Liu XY, Dai X, Liu YH, Liu SJ (2008) Paracoccus halophilus sp. nov., isolated from marine sediment of the South China Sea, China, and emended description of genus Paracoccus Davis 1969. Int J Syst Evol Microbiol 58:257–261
Ludwig W, Mittenhuber G, Friedrich CG (1993) Transfer of Thiosphaera pantotropha to Paracoccus denitrificans. Int J Syst Bacteriol 43:363–367
Lyu L, Zhi B, Lai Q, Shao Z, Yu Z (2020) Paracoccus xiamenensis sp. nov., isolated from seawater on the Xiamen. Int J Syst Evol Microbiol 70:4285–4290
Meng XL, Ming H, Huang JR, Zhang LY, Cheng LJ, Zhao ZL, Ji WL, Li WJ, Nie GX (2018) Paracoccus halotolerans sp. nov., isolated from a salt lake. Int J Syst Evol Microbiol 69:523–528
Minnikin D, O’Donnell A, Goodfellow M, Alderson G, Athalye M, Schaal A, Parlett JH (1984) An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Appl Bacteriol 2:233–241
Mitchell AL, Attwood TK, Babbitt PC (2019) InterPro in 2019: improving coverage, classification and access to protein sequence annotations. Nucleic Acids Res 47:D351–D360
Mu DS, Liang QY, Wang XM, Lu DC, Shi MJ, Chen GJ, Du ZJ (2018) Metatranscriptomic and comparative genomic insights into resuscitation mechanisms during enrichment culturing. Microbiome 6:230
Qin QL, Xie BB, Zhang XY, Chen XL, Zhou BC, Zhou J, Oren A, Zhang YZ (2014) A proposed genus boundary for the prokaryotes based on genomic insights. J Bacteriol 196:2210–2215
Richter M, Rosselló-Móra R (2009) Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci 106:19126–19131 (USA)
Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids. MIDI Tech Note 101:1–6
Sheu SY, Hsieh TY, Young CC, Chen WM (2018) Paracoccus fontiphilus sp. nov., isolated from a freshwater spring. Int J Syst Evol Microbiol 68:2054–2060
Smibert RM, Krieg NR (1994) Phenotypic characterization. In: Gerhardt P, Murray RGE, Wood WA, Krieg NR (eds) Methods for general and molecular bacteriology. American Society for Microbiology, Washington, DC, pp 607–654
Xu XW, Wu YH, Wang CS, Oren A, Zhou PJ, Wu M (2007) Haloferax larsenii sp. nov., an extremely halophilic archaeon from a solar saltern. Int J Syst Evol Microbiol 57:717–720
Xue H, Piao CG, Guo MW, Wang LF, Li Y (2017) Paracoccus aerius sp. nov. isolated from air. Int J Syst Evol Microbiol 67:2586–2591
Yoon SH, Ha SM, Kwon S, Lim J, Kim Y, Seo H, Chun J (2017) Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol 67:1613–1617
Zhang G, Jiao K, Xie F, Pei S, Jiang L (2019) Paracoccus subflavus sp. nov., isolated from Pacific Ocean sediment. Int J Syst Evol Microbiol 69:1472–1476
Acknowledgements
This work was supported by the National Natural Science Foundation of China (32070111, 31870105). We thank Dr. Zong-Jun Du (Marine College, Shandong University, Weihai, 264209, PR China) for kindly providing the strain Paracoccus denitrificans DSM 413T.
Author information
Authors and Affiliations
Contributions
WF, MD and LQ designed research and project outline. LYL, GXF, FXY and HZS performed isolation, deposition and identification. WF, GPF, DZJ and LQ drafted the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Human and animal rights
This article does not contain any studies with animals performed by any of the authors.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Communicated by Erko Stackebrandt.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, F., Gong, XF., Meng, D. et al. Paracoccus binzhouensis sp. nov., isolated from activated sludge. Arch Microbiol 203, 3007–3013 (2021). https://doi.org/10.1007/s00203-021-02286-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-021-02286-7