Abstract
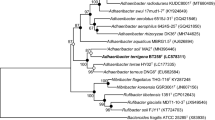
A novel bacterial strain, designated ZR32T, was isolated from briquette warehouse soil in Ulsan (Korea). The strain was aerobic, showing pink-colored colonies on R2A agar. Phylogenetic analyses based on 16S rRNA gene sequences showed that strain ZR32T was closely related to Mucilaginibacter soli R9-65T (97.0%), Mucilaginibacter gynuensis YC7003T (96.9%), and Mucilaginibacter lutimaris BR-3T (96.8%). The values of DNA–DNA relatedness related two highest strains M. soli R9-65T and M. gynuensis YC7003T were 31.2 ± 6.9% and 19.7 ± 0.3%, respectively. Its genome size was 3.9 Mb, comprising 3402 predicted genes. The DNA G+C content of strain ZR32T was 43.0 mol%. The major cellular fatty acids (> 5% of total) were summed feature 3 (C16:1ω6c and/or C16:1ω7c), C16:0, C16:1ω5c, iso-C15:0, iso-C17:0 3-OH, and C17:1ω9c. The major respiratory quinine was menaquinone-7 (MK-7). The major polar lipids were phosphatidylethanolamine, two unidentified phospholipids, one unidentified sphingolipid, and one unidentified polar lipid. Strain ZR32T showed distinctive characteristics such as the temperature and pH for growth ranges, being positive for β-glucosidase, salicin production, negative for N-acetyl-glucosamine assimilation, being resistant to carbenicillin and piperacillin to related species. On the basis of phenotypic, chemotaxonomic, and phylogenetic data, strain ZR32T represents a novel species of the genus Mucilaginibacter, for which the name Mucilaginibacter hurinus sp. nov. is proposed. The type strain is ZR32T (= KCTC 62193 = CCTCC AB 2017285).

Similar content being viewed by others
Abbreviations
- KCTC:
-
Korean Collection for Type Cultures
- NRRL:
-
Agricultural Research Service Culture Collection
- CCTCC:
-
China Center for Type Culture Collection
- KACC:
-
Korean Agricultural Culture Collection
- TEM:
-
Transmission electron microscopy
- MK-7:
-
Menaquinone-7
- MK-6:
-
Menaquinone-6
- NCBI:
-
National Center for Biotechnology Information
- LB:
-
Luria–Bertani
- NA:
-
Nutrient agar
- TSA:
-
Trypticase soy agar
- MH:
-
Mueller–Hinton
- MEGA:
-
Molecular evolutionary genetics analysis
- GDDCM:
-
Guangdong Detection Center of Microbiology
References
Aydogan EL, Busse HJ, Moser G, Müller C, Kämpfer P et al (2017) Proposal of Mucilaginibacter galii sp. nov., isolated from leaves of Galium album. Int J syst Evol Microbiol 67:1318–1326. https://doi.org/10.1099/ijsem.0.001808
Baik KS, Park SC, Kim EM, Lim CH, Seong CN (2010) Mucilaginibacter rigui sp. nov., isolated from wetland freshwater, and emended description of the genus Mucilaginibacter. Int J Syst Evol Microbiol 60:134–139. https://doi.org/10.1099/ijs.0.011130-0
Bernardet JF, Nakagawa Y, Holmes B, Subcommittee on the taxonomy of Flavobacterium, and Cytophaga-like bacteria of the International Committee on Systematics of Prokaryotes (2002) Proposed minimal standards for describing new taxa of the family Flavobacteriaceae and emended description of the family. Int J Syst Evol Microbiol 52:1049–1070. https://doi.org/10.1099/00207713-52-3-1049
Cappuccino JG, Sherman N (2002) Microbiology: a laboratory manual, 6th edn. Benjamin/Cummings, Menlo Park
Chen XY, Zhao R, Tian Y, Kong BH, Li XD et al (2014) Mucilaginibacter polytrichastri sp. nov., isolated from a moss (Polytrichastrum formosum), and emended description of the genus Mucilaginibacter. Int J Syst Evol Microbiol 64:1395–1400. https://doi.org/10.1099/ijs.0.055335-0
Chen WM, Hsieh TY, Sheu SY (2018) Mucilaginibacter amnicola sp. nov., isolated from a freshwater creek. Int J syst Evol Microbiol 68:394–401. https://doi.org/10.1099/ijsem.0.002518
Chun J, Oren A, Ventosa A, Christensen H, Arahal DR et al (2018) Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int J Syst Evol Microbiol 68(1):461–466. https://doi.org/10.1099/ijsem.0.002516
Collins MD, Jones D (1980) Lipids in the classification and identification of coryneform bacteria containing peptidoglycans based on 2,4-diaminobutyric acid. J Appl Bacteriol 48:459–470. https://doi.org/10.1111/j.1365-2672.1980.tb01036.x
Dong XZ, Cai MY (2001) Determinative manual for routine bacteriology. Scientific Press, Beijing
Ezaki T, Hashimoto Y, Yabuuchi E (1989) Fluorometric deoxyribonucleic acid-deoxyribonucleic acid hybridization in microdilution wells as an alternative to membrane filter hybridization in which radioisotopes are used to determine genetic relatedness among bacterial strains. Int J Syst Bacteriol 39:224–229. https://doi.org/10.1099/00207713-39-3-224
Fan H, Su C, Wang Y, Yao J, Zhao K (2008) Sedimentary arsenite-oxidizing and arsenate-reducing bacteria associated with high arsenic groundwater from Shanyin Northwestern China. J Appl Microbiol 105:529–539. https://doi.org/10.1111/j.1365-2672.2008.03790.x
Fan X, Tang J, Nie L, Environmental Microbiome et al (2018) High-quality-draft genome sequence of the heavy metal resistant and exopolysaccharides producing bacterium Mucilaginibacter pedocola TBZ30T. Stand Genom Sci 13:34. https://doi.org/10.1186/s40793-018-0337-8
Felsenstein J (1981) Evolutionary trees from DNA sequence: a maximum likelihood approach. J Mol Evol 17:368–376. https://doi.org/10.1007/BF01734359
Felsenstein JP (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Han SI, Lee HJ, Lee HR, Kim KK, Whang KS (2012) Mucilaginibacter polysacchareus sp. nov., an exopolysaccharideproducing bacterial species isolated from the rhizoplane of the herb Angelica sinesis. Int J Syst Evol Microbiol 62:632–637. https://doi.org/10.1099/ijs.0.029793-0
Hwang YM, Baik KS, Seong CN (2014) Mucilaginibacter defluvii sp. nov., isolated from a dye wastewater treatment facility. Int J syst Evol Microbiol 64:565–571. https://doi.org/10.1099/ijs.0.055590-0
Jiang F, Dai J, Wang Y, Xue X, Xu M et al (2012) Mucilaginibacter soli sp. nov., isolated from Arctic tundra soil. Int J Syst Evol Microbiol 62:1630–1635. https://doi.org/10.1099/ijs.0.033902-0
Kämpfer P, Busse HJ, Mclnroy JA, Glaeser SP (2014) Mucilaginibacter auburnensis sp. nov., isolated from a plant stem. Int J Syst Evol Microbiol 64:1736–1742. https://doi.org/10.1099/ijs.0.060848-0
Khan HJ, Chung EJ, Jeon CO, Chung YR (2013) Mucilaginibacter gynuensis sp. nov., isolated from rotten wood. Int J syst Evol Microbiol 63:3225–3231. https://doi.org/10.1099/ijs.0.050153-0
Kim JH, Kang SJ, Jung YT, Oh K, Yoon JH (2012a) Mucilaginibacter lutimaris sp. nov., isolated from a tidal flat sediment. Int J syst Evol Microbiol 62:515–519. https://doi.org/10.1099/ijs.0.030213-0
Kim OS, Cho YJ, Lee K, Yoon SH, Kim M et al (2012b) Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol 62:716–721. https://doi.org/10.1099/ijs.0.038075-0
Kim MM, Siddiqi MZ, Im WT (2017) Mucilaginibacter ginwenosidivorans sp. nov., isolated from soil of ginseng field. Curr Microiol 74(12):1382–1388. https://doi.org/10.1007/s00284-017-1329-4#Bib1
Kimura MJ (1980) A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16(2):111–120
Krogh A, Larsson B, Heijin GV, Sonnhammer ELL (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305:567–580
Kroppenstedt RM (1985) Fatty acid and menaquinone analysis of actinomycetes and related organisms. In: Goodfellow M, Minnikin DE (eds) Chemical methods in bacterial systematics. Academic Press, pp 173–199. https://www.mendeley.com/research-papers/fatty-acid-menaquinone-analysis-actinomycetes-related-organisms/authors/
Lee HR, Han SI, Rhee KH, Whang KS (2013) Mucilaginibacter herbaticus sp. nov., isolated from the rhizosphere of the medicinal plant Angelicasinensis. Int J Syst Evol Microbiol 63:2787–2793. https://doi.org/10.1099/ijs.0.038398-0
Lee KC, Kim KK, Eom MK, Kim JS, Kim DS et al (2017) Mucilaginibacter craterilacus sp. nov., isolated from sediment soil of a crater lake. Int J Syst Evol Microbiol 67:2891–2896. https://doi.org/10.1099/ijsem.0.002043
Lee SY, Siddiqi MZ, Kim SY, Yu HS, Lee JH et al (2018) Mucilaginibacter panaciglaebae sp. nov., isolated from soil of a ginseng field. Int J Syst Evol Microbiol 68:149–154. https://doi.org/10.1099/ijsem.0.002473
Li R, Zhu H, Ruan J, Qian W, Fang X et al (2010) De novo assembly of human genomes with massively parallel short read sequencing. Genome Res 20(2):265–272
Liu Q, Siddiqi MZ, Kim MS, Kim SY, Im WT (2017) Mucilaginibacter hankyongensis sp. nov., isolated from soil of ginseng field Baekdu Mountain. J Microbiol 55(7):525–530. https://doi.org/10.1007/s12275-017-7180-2
Luo JH, Liu BH, Xie YL, Li ZY, Huang WH et al (2012) SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 1(1):18. https://doi.org/10.1186/2F2047-217X-1-18
Madhaiyan M, Poonguzhali S, Lee JS, Senthilkumar M, Lee KC et al (2010) Mucilaginibacter gossypii sp. and Mucilaginibacter gossypiicola sp. nov., plant-growth-promoting bacteria isolated from cotton rhizosphere soils. Int J syst Evol Microbiol 60:2451–2357. https://www.microbiologyresearch.org/docserver/fulltext/ijsem/60/10/2451.pdf?expires=1568098026&id=id&accname=guest&checksum=618CFD2997ADBE0F19B6D9339FE8F45F
Männistö MK, Tiirola M, McConnell J, Häggblom MM (2010) Mucilaginibacter frigoritolerans sp. nov., Mucilaginibacter lappiensis sp. nov. and Mucilaginibacter mallensis sp. nov., isolated from soil and lichen samples. Int J Syst Evol Microbiol 60:2849–2856. https://doi.org/10.1099/ijs.0.019364-0
Morgan NP, Paramvir SD, Adam PA (2009) FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol 26(7):1641–1650
Paiva G, Abreu P, Proenca DN, Santos S, Nobre MF et al (2014) Mucilaginibacter pineti sp. nov., isolated from Pinus pinaster wood from a mixed grove of pines trees. Int J Syst Evol Microbiol 64:2223–2228. https://doi.org/10.1099/ijs.0.057737-0
Pankratov TA, Tindall BJ, Liesack W, Dedysh SN (2007) Mucilaginibacter paludis gen. nov., sp. nov. and Mucilaginibacter gracilis sp. nov., pectin-, xylan- and laminarin-degrading members of the family Sphingobacteriaceae from acidic Sphagnum peat bog. Int J Syst Evol Microbiol 57:2349–2354. https://doi.org/10.1099/ijs.0.65100-0
Petersen TN, Brunak S, Heijine GV, Nielsen H (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785–786
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4(4):406–425
Sasser M (2001) Identification of bacteria by gas chromatography of cellular fatty acids. MIDI Technical Note 101 DE: MIDI Inc, Newark
Sedláček I, Pantůček R, Králová S, Mašlaňová S, Holochová P et al (2017) Mucilaginibacter terrae sp. nov., isolated from Antarctic soil. Int J syst Evol Microbiol 67:4002–4007. https://doi.org/10.1099/ijsem.0.002240
Smibert RM, Krieg NR (1994) Phenotypic characterization. In: Methods for general and molecular bacteriology. American Society for Microbiology, Washington, DC, pp 607–654. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC205328/pdf/jbacter00026-0002.pdf
Tamaoka J, Komagata K (1994) Determination of DNA base composition by reversed-phase high-performance liquid chromatography. FEMS Microbiol Lett 25:125–128
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Tang J, Huang J, Qiao Z, Wang R, Wang G (2016) Mucilaginibacter pedocola sp. nov., isolated from a heavy-metal-contaminated paddy field. Int J Syst Evol Microbiol 66:1–6. https://doi.org/10.1099/ijsem.0.001306
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Urai M, Aizawa T, Nakagawa Y, Nakajima M, Sunairi M (2008) Mucilaginibacter kameinonensis sp., nov., isolated from garden soil. Int J Syst Evol Microbiol 58:2046–2050. https://doi.org/10.1099/ijs.0.65777-0
Wei JC, Sun LN, Yuan ZX, Hou XT, Yang ED et al (2017) Mucilaginibacter rubeus sp. nov., isolated from rhizosphere soil. Int J Syst Evol Microbiol 67:3099–3104. https://doi.org/10.1099/ijsem.0.002101
Wu S, Zhu Z, Fu L, Niu B, Li W (2011) WebMGA: a customizable web server for fast metagenomic sequence analysis. BMC Genom 12:444. https://doi.org/10.1186/1471-2164-12-444
Yoon JH, Kang SJ, Park S, Oh TK (2012) Mucilaginibacter litoreus sp. nov., isolated from marine sand. Int J Syst Evol Microbiol 62:2822–2877. https://doi.org/10.1099/ijs.0.034900-0
Acknowledgements
This work was supported by the Development Program of China (National Development and Reform Commission; 2016YFD0800702). The authors are grateful to Prof. Dr. Bernhard Schink for the Latin construction of the species name. We acknowledge the Guangdong Detection Center of Microbiology (GDDCM) for fatty acid composition, polar lipids, and the DNA–DNA hybridization analyses. In addition, Genome was analyzed at Wuhan Frasergen Bioinformatics Co., Ltd.
Author information
Authors and Affiliations
Contributions
LC: she has brought soil sample from Korea. She supported the procedures for the characterization for the strain ZR32T and the closest type strains. She conducted the PCR and sequencing of the 16S rRNA gene. She had constructed the phylogenetic trees, and Gram staining of the cells. She determined the cellular fatty acids. She had prepared the lyophilized powder of strain ZR32T and the closest related strains for analysis of polar lipids, isoprenoid quinone and DNA–DNA hybridization. She had conducted the experiments for bacterial phenotypic characteristics using the API strips. She had performed the analysis of the genome of ZR32T. She also made the tables and figure and wrote the manuscript. XZ: She helped Ms. Choi to collect colonies. In addition, she had conducted the experiments for bacterial phenotypic characteristics (growth temperature test, growth pH test, growth NaCl test, and motility test). YS: she supported the whole methods for the characterization for the strain ZR32T and the closest type strains. MW: he helped to correct grammar errors. In addition, he helped to revise the paper. GW: she supported the funding for the study. In addition, she advised about the way of experiments. ML: she proposed strain ZR32T as a new species. In addition, she advised about experimental skills.
Corresponding author
Ethics declarations
Conflict of interest
There is no conflict of interest.
Additional information
Communicated by Erko Stackebrandt.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The GenBank/EMBL/DDBJ accession number for the 16S rRNA gene sequence of Mucilaginibacter hurinus ZR32T is MF919638. Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the accession QGDC00000000.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Choi, L., Zhao, X., Song, Y. et al. Mucilaginibacter hurinus sp. nov., isolated from briquette warehouse soil. Arch Microbiol 202, 127–134 (2020). https://doi.org/10.1007/s00203-019-01720-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-019-01720-1