Abstract
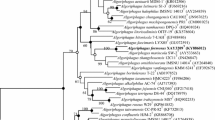
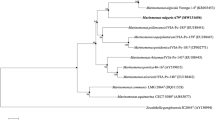
A Gram-staining-negative, strictly aerobic, non-motile, ovoid- to rod-shaped bacterium, designated as HZ20T, was isolated from the surface of a brown seaweed (Laminaria japonica) sample collected from the East China Sea. Colonies are 1.0–2.0 mm in diameter, smooth, circular, convex and yellow after grown on MA at 28 °C for 72 h. The strain was found to grow at 4–50 °C (optimum, 37 °C), pH 5.0–9.5 (optimum, pH 7.0–7.5) and with 0–10% (w/v) NaCl (optimum, 1.0–1.5%). Chemotaxonomic analysis showed ubiquinone-8 as the only quinone, C17:0 cyclo, C16:0, summed feature 8 (C18:1ω7c and/or C18:1ω6c) and summed feature 2 (C12:0 aldehyde/unknown 10.9525/C16:1 iso I/C14:0 3OH) as the major fatty acids (> 5%), and diphosphatidylglycerol, phosphatidylethanolamine, phosphatidylglycerol, one unidentified amino phospholipid, two unidentified phospholipids, five unidentified glycolipid and two unidentified lipids as the polar lipids. The DNA G + C content was 55.5 mol %. 16S rRNA gene sequences of the isolate showed highest similarities to Bordetella flabilis AU10664T (97.1%), Parapusillimonas granuli Ch07T (97.1%), Paracandidimonas soli IMT-305T (97.1%), Kerstersia gyiorum LMG5906T (97.0%) and Bordetella sputigena LMG 28641T (97.0%). The phylogenetic trees using 16S rRNA gene and genome sequences both showed that the strain HZ20T formed a deep branch separated from other related genera, indicating that it represents a novel species of a novel genus. The calculated average nucleotide identity (ANI) and percent of conserved proteins (POCP) values using genome sequences of strain HZ20T and related strains also support this conclusion. Based on the phenotypic properties and phylogenetic distinctiveness, we propose strain HZ20T (= MCCC 1K03465T = KCTC 62330T) to represent a novel species of a novel genus with the name Algicoccus marinus gen. nov. sp. nov.

Similar content being viewed by others
Abbreviations
- POCP:
-
Percent of conserved proteins
- ANI:
-
Average nucleotide identity
- DPG:
-
Diphosphatidylglycerol
- PG:
-
Phosphatidylglycerol
- PE:
-
Phosphatidylethanolamine
- PS:
-
Phosphatidylserine
- APL:
-
Unidentified aminophospholipid
- PL:
-
Unidentified phospholipid
- AL:
-
Unidentified aminolipid
References
Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O (2008) The RAST server: Rapid annotations using subsystems technology. BMC Genom 9:75
Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T (2009) trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25:1972–1973
Chun J, Oren A, Ventosa A, Christensen H, Arahal DR, Da Costa MS, Rooney AP, Yi H, Xu XW, Meyer SD, Trujillo ME (2018) Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int J Syst Evol Microbiol 68:461–466
Coenye T, Vancanneyt M, Cnockaert MC, Falsen E, Swings J, Vandamme P (2003) Kerstersia gyiorum gen. nov., sp. nov., a novel Alcaligenes faecalis-like organism isolated from human clinical samples, and reclassification of Alcaligenes denitrificans Ruger and Tan 1983 as Achromobacter denitrificans comb. nov. Int J Syst Evol Microbiol 53(6):1825–1831
Coenye T, Vancanneyt M, Falsen E, Swings J, Vandamme P (2013) Achromobacter insolitus sp. nov. and Achromobacter spanius sp. nov., from human clinical samples. Int J Syst Evol Microbiol 53:1819–1824
Felsenstein J (1981) Evolutionary trees from DNA-sequences—a maximum-likelihood approach. J Mol Evol 17(6):368–376
Fischer S, Brunk BP, Chen F, Gao X, Harb OS, Iodice JB, Shanmugam D, Roos DS, Stoeckert CJ (2011) Using OrthoMCL to assign proteins to OrthoMCL-DB groups or to cluster proteomes into new Ortholog groups. Curr Protoc Bioinform 6(12):1–19
Gomila M, Tvrzová L, Teshim A, Sedláček I, González EN, Zdráhal Z, Šedo O, González JF, Bennasar A, Moore ERB, Lalucat J, Murialdo SE (2011) Achromobacter marplatensis sp. nov., isolated from a pentachlorophenol-contaminated soil. Int J Syst Evol Microbiol 61:2231–2237
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780
Keampfer P, Busse HJ, McInroy JA, Glaeser SP (2017) Paracandidimonas soli gen. nov., sp. nov., isolated from soil. Int J Syst Evol Microbiol 67:1740–1745
Kersters K, Hinz KH, Hertle A, Segers P, Lievens A, Siegmann O, De Ley J (1984) Bordetella avium sp. nov., isolated from the respiratory tracts of turkeys and other birds. Int J Syst Bacteriol 34:56–70
Kim YJ, Kim MK, Im WT, Srinivasan S, Yang DC (2010) Parapusillimonas granuli gen. nov., sp. nov., isolated from granules from a wastewater-treatment bioreactor. Int J Syst Evol Microbiol 60:1401–1406
Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H, Park SC, Jeon YS, Lee JH, Yi H, Won S, Chun J (2012) Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol 62:716–721
Komagata K, Suzuki K (1987) Lipid and cell-wall analysis in bacterial systematics. Method Microbiol 19:161–207
Lagesen K, Hallin P, Rodland EA, Staerfeldt HH, Rognes T, Ussery DW (2007) RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res 35(9):3100–3108
Lee I, Kim YO, Park SC, Chun J (2016) OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int J Syst Evol Microbiol 66:1100–1103
Leifson E (1963) Determination of carbohydrate metabolism of marine bacteria. J Bacteriol 85:1183–1184
Minnikin DE, Odonnell AG, Goodfellow M, Alderson G, Athalye M, Schaal A, Parlett JH (1984) An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Meth 2(5):233–241
Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ (2014) IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32:268–274
Parte AC (2018) LPSN—list of prokaryotic names with standing in nomenclature (bacterio.net), 20 years on. Int J Syst Evol Microbiol 68(6):1825–1829
Qin QL, Xie BB, Zhang XY, Chen XL, Zhou BC, Zhou JZ, Oren A, Zhang YZ (2014) A proposed genus boundary for the prokaryotes based on genomic insights. J Bacteriol 196(12):2210–2215
Saitou N, Nei M (1987) The neighbor-joining method—a new method for reconstructing phylogenetic trees. Mol Biol Evol 4(4):406–425
Su Y, Han SB, Wang RJ, Yu XY, Fu GY, Chen C, Zhang CY, Li Y, Sun C, Wu M (2017) Microbaculum marinum gen. nov., sp. nov., isolated from deep seawater. Int J Syst Evol Microbiol 67(4):812–817
Sun C, Wang RJ, Su Y, Fu GY, Zhao Z, Yu XY, Zhang CY, Chen C, Han SB, Huang MM, Lv ZB, Wu M (2017) Hyphobacterium vulgare gen. nov., sp. nov., a novel alphaproteobacterium isolated from seawater. Int J Syst Evol Microbiol 67(5):1169–1176
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28(10):2731–2739
Tazato N, Handa Y, Nishijima M, Kigawa R, Sano C, Sugiyama J (2015) Novel environmental species isolated from the plaster wall surface of mural paintings in the Takamatsuzuka tumulus: Bordetella muralis sp. nov., Bordetella tumulicola sp. nov. and Bordetella tumbae sp. nov. Int J Syst Evol Microbiol 65:4830–4838
Vandamme P, Hommez J, Vancanneyt M, Monsieurs M, Hoste B, Cookson B, Wirsing Von König CH, Kersters K, Blackall PJ (1995) Bordetella hinzii sp. nov., isolated from poultry and humans. Int J Syst Bacteriol 45:37–45
Vandamme P, Heyndrickx M, Vancanneyt M, Hoste B, De Vos P, Falsen E, Kersters K, Hinz KH (1996) Bordetella trematum sp. nov., isolated from wounds and ear infections in humans, and reassessment of Alcaligenes denitrificans Rüger and Tan 1983. Int J Syst Bacteriol 46:849–858
Vandamme P, De Brandt E, Houf K, De Baere T (2012) Kerstersia similis sp. nov., isolated from human clinical samples. Int J Syst Evol Microbiol 62(9):2156–2159
Vandamme P, Moore ERB, Cnockaert M, Peeters C, Svensson-Stadler L, Houf K, Spilker T, Lipuma JJ (2013a) Classification of Achromobacter genogroups 2, 5, 7 and 14 as Achromobacter insuavis sp. nov., Achromobacter aegrifaciens sp. nov., Achromobacter anxifer sp. nov. and Achromobacter dolens sp. nov., respectively. Syst Appl Microbiol 36:474–482
Vandamme P, Moore ERB, Cnockaert M, De Brandt E, Svensson-Stadler L, Houf K, Spilker T, Lipuma JJ (2013b) Achromobacter animicus sp. nov., Achromobacter mucicolens sp. nov., Achromobacter pulmonis sp. nov. and Achromobacter spiritinus sp. nov., from human clinical samples. Syst Appl Microbiol 36:1–10
Vandamme P, Peeters C, Cnockaert M, Inganas E, Falsen E, Moore ER, Nunes OC, Manaia CM, Spilker T, Lipuma JJ (2015) Bordetella bronchialis sp. nov., Bordetella flabilis sp. nov. and Bordetella sputigena sp. nov., isolated from human respiratory specimens, and reclassification of Achromobacter sediminum Zhang et al. 2014 as Verticia sediminum gen. nov., comb. nov. Int J Syst Evol Microbiol 65:3674–3682
Vandamme P, Peeters C, Inganas E, Cnockaert M, Houf K, Spilker T, Moore ERB, Lipuma JJ (2016) Taxonomic dissection of Achromobacter denitrificans Coenye et al. 2003 and proposal of Achromobacter agilis sp. nov., nom. rev., Achromobacter pestifer sp. nov., nom. rev., Achromobacter kerstersii sp. nov. and Achromobacter deleyi sp. nov. Int J Syst Evol Microbiol 66:3708–3717
Von Wintzingerode F, Schattke A, Siddiqui RA, Rösick U, Göbel UB, Gross R (2001) Bordetella petrii sp. nov., isolated from an anaerobic bioreactor, and emended description of the genus Bordetella. Int J Syst Evol Microbiol 51:1257–1265
Weyant RS, Hollis DG, Weaver RE, Amin MFM, Steigerwalt AG, O’connor SP, Whitney AM, Daneshvar MI, Moss CW, Brenner DJ (1995) Bordetella holmesii sp. nov., a new gram-negative species associated with septicemia. J Clin Microbiol 33:1–7
Yabuuchi E, Yano I (1981) Achromobacter gen. nov. and Achromobacter xylosoxidans (ex Yabuuchi and Ohyama 1971) nom. rev. Int J Syst Bacteriol 31:477–478
Yabuuchi E, Kawamura Y, Kosako Y, Ezaki T (1998) Emendation of the genus Achromobacter and Achromobacter xylosoxidans (Yabuuchi and Yano) and proposal of Achromobacter ruhlandii (Packer and Vishniac) comb. nov., Achromobacter piechaudii (Kiredjian et al.) comb. nov., and Achromobacter xylosoxidans subsp. denitrificans (Rüger and Tan) comb. nov. Microbiol Immunol 42:429–438
Zhang XQ, Wu YH, Zhou X, Zhang X, Xu XW, Wu M (2016) Parvularcula flava sp. nov., an alphaproteobacterium isolated from surface seawater of the South China Sea. Int J Syst Evol Microbiol 66(9):3498–3502
Zhang WY, Yuan Y, Su DQ, He XP, Han SB, Epstein SS, He S, Wu M (2018a) Gallaecimonas mangrovi sp. nov., a novel bacterium isolated from mangrove sediment. Antonie van Leeuwenhoek 111(10):1855–1862
Zhang WY, Yuan Y, Su DQ, Ding LJ, Yan XJ, Wu M, Epstein SS, He S (2018b) Saccharospirillum mangrovi sp. nov., a bacterium isolated from mangrove sediment. Int J Syst Evol Microbiol 68(9):2813–2818
Acknowledgements
This work is supported by the Zhejiang Provincial Natural Science Foundation (LQ19C010005) and the Science Foundation of Zhejiang Sci-Tech University (16042186-Y).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare that there are no conflicts of interest. Informed consent was obtained from all the individual participants included in the study.
Additional information
Communicated by Erko Stackebrandt.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The GenBank/EMBL/DDBJ accession numbers for the 16S rRNA gene sequences and complete genome sequences of strain HZ20T are MG712864 and CP028901, respectively. The DPD number of strain HZ20T is TA00577.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ying, JJ., Zhang, SL., Huang, CY. et al. Algicoccus marinus gen. nov. sp. nov., a marine bacterium isolated from the surface of brown seaweed Laminaria japonica. Arch Microbiol 201, 943–950 (2019). https://doi.org/10.1007/s00203-019-01664-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-019-01664-6